Comparing LFQ and Isobaric Labeling Strategies in Proteomics Quantification
- Label-Free Quantitation Service
- TMT/iTRAQ Labeling-Based Quantitative Service
- Quantitative Proteomics Analysis Service
- Isotope Labeling-Based Quantitative Service
Quantitative proteomics plays a critical role in modern life science research, offering a window into the dynamic regulation of cellular systems under physiological and pathological conditions. Unlike protein identification, which focuses solely on cataloging protein species, protein quantification aims to measure changes in abundance, enabling researchers to investigate differential expression, post-translational modifications, and signaling responses across experimental conditions.
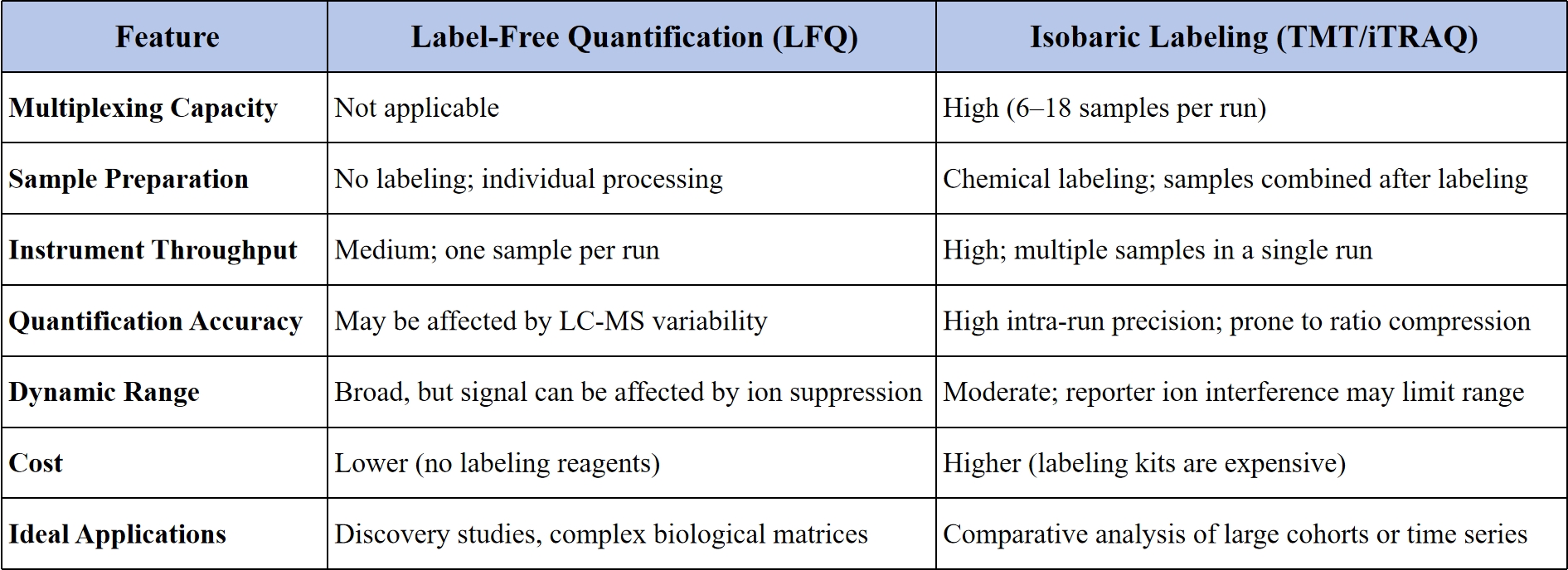
Two of the most widely adopted mass spectrometry (MS)-based quantification strategies are label-free quantification (LFQ) and isobaric labeling methods, such as tandem mass tags (TMT) and isobaric tags for relative and absolute quantitation (iTRAQ). Each method brings unique advantages and challenges depending on study design, sample type, and analytical goals.
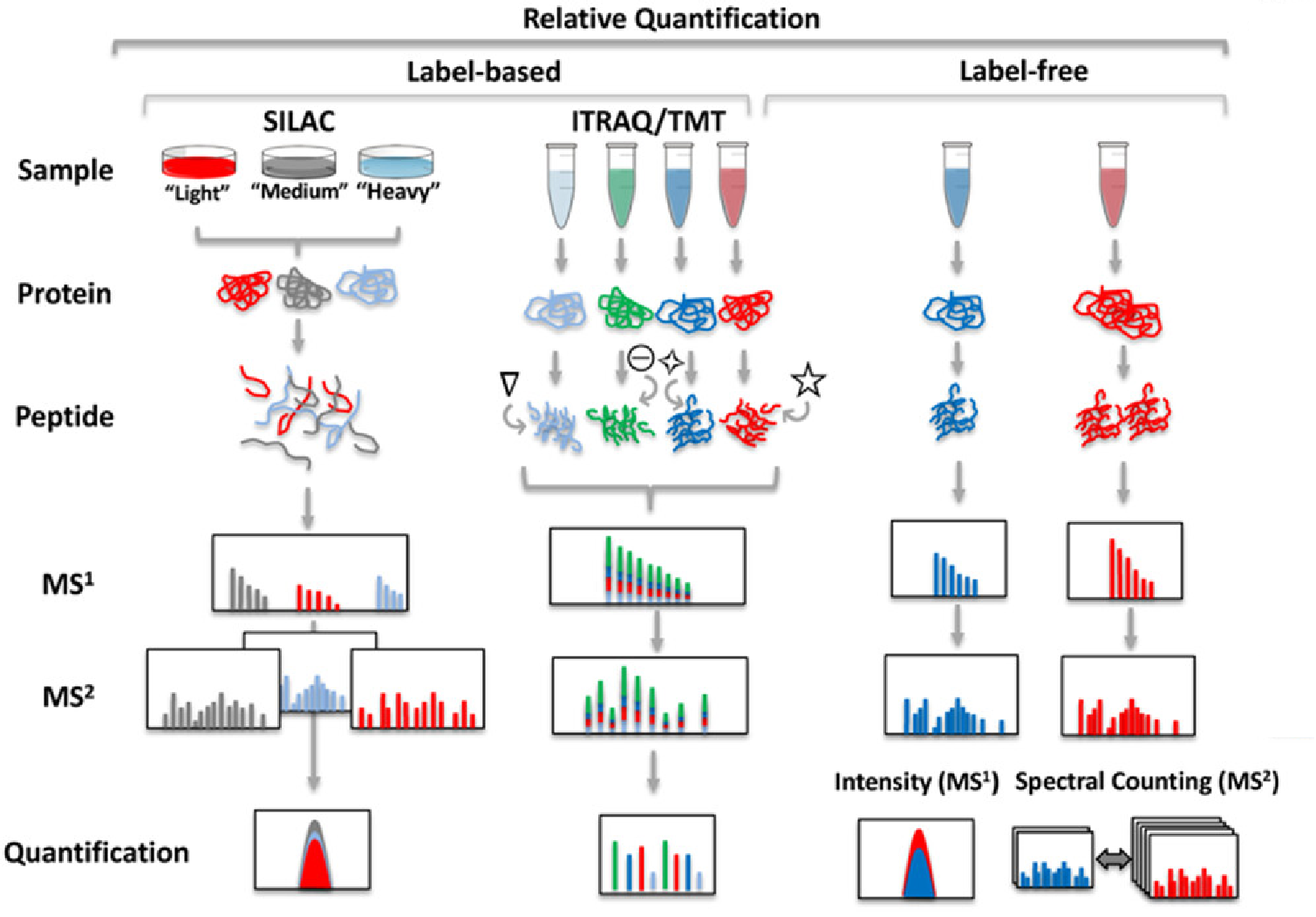
Calvete, JJ. et al. Mass Spectrom Rev. 2024.
Figure1. Generalized Comparison of Different Quantitative Proteomic Strategies
Select Service
Label Free Quantification (LFQ)
Label-free quantification is a widely used strategy that estimates protein abundance directly from the raw signal intensity of peptide ions or from spectral counting during LC-MS analysis. This method relies on the assumption that the ion signal of a peptide is proportional to its concentration in the sample. In typical LFQ workflows, proteins are enzymatically digested into peptides, which are then separated by liquid chromatography and detected by high-resolution mass spectrometry. Quantitative comparisons across samples are achieved through software-based alignment of retention times and normalization of signal intensities.
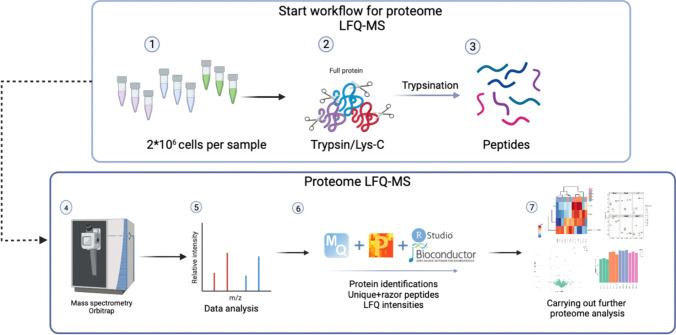
Sulaj E, et al, 2024.
Figure 2. Workflow of Label-free Quantification
Advantages:
✅No chemical labeling requiredLFQ does not rely on external tags, eliminating potential variability introduced by incomplete or inconsistent labeling reactions.
✅Cost-effective for large sample setsSince no labeling reagents are needed, LFQ is budget-friendly, especially for exploratory studies with high sample throughput.
✅High sample flexibilityLFQ is compatible with a wide range of biological materials—including tissues, body fluids, and FFPE samples—without the need for specialized preparation.
✅ Broad dynamic rangeLFQ can detect changes across a wide range of protein abundances, making it well-suited for complex biological samples.
Because LFQ does not require chemical or metabolic labeling, it is cost-effective and flexible—particularly suitable for studies with large numbers of samples or when labeling is not feasible, such as in clinical tissues or body fluids. However, it is sensitive to run-to-run variability and often requires robust normalization and statistical processing to ensure quantitative accuracy.
Isobaric Labeling (TMT/iTRAQ)
Isobaric labeling methods, including TMT and iTRAQ, involve covalently tagging peptides from different samples with chemically identical labels that differ only in isotopic composition. These tags produce identical precursor masses but release reporter ions of distinct masses during MS/MS fragmentation. After labeling, samples are combined and analyzed together in a single LC-MS/MS run, allowing simultaneous quantification of multiple conditions.
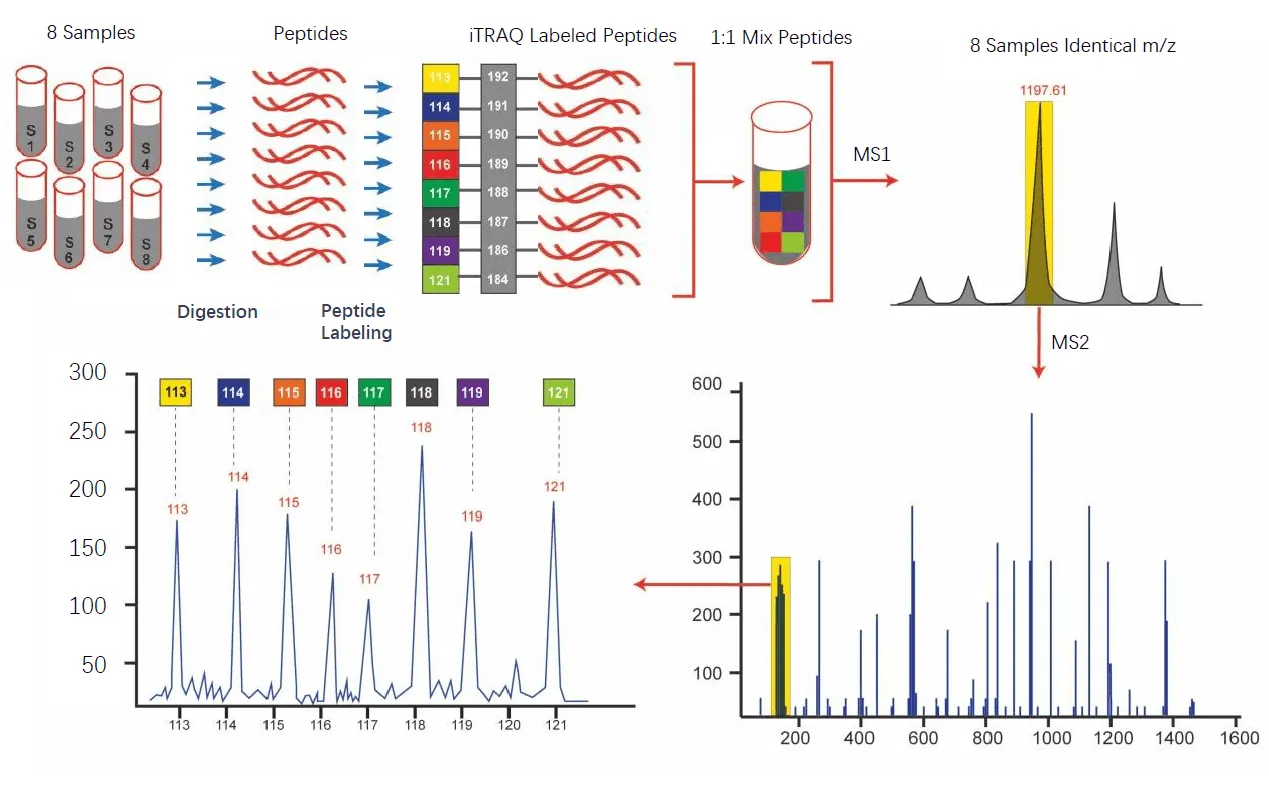
Figure 3. Workflow of TMT Quantitative Proteomics
Advantages:
✅High-throughput multiplexing
Isobaric labeling allows simultaneous analysis of 6 to 18 samples in a single LC-MS run, significantly increasing experimental efficiency.
✅Improved data consistency
Since all labeled samples are analyzed together, inter-sample variability is minimized—ideal for time-course or multi-group comparisons.
✅Enhanced quantification precision
Quantitative comparisons are performed within the same run, reducing technical variation and increasing confidence in differential expression results.
✅Ideal for complex experimental designs
Isobaric labeling is particularly useful for large-scale studies involving multiple biological conditions, treatment groups, or replicates.
A major advantage of isobaric labeling is its ability to multiplex—enabling 6 to 18 samples (depending on the reagent version) to be analyzed in one experiment with high precision and minimal technical variability. This is particularly useful for studies requiring tight inter-sample comparability, such as time-course experiments or patient cohort analysis. However, quantification at the MS2 level is susceptible to ratio distortion due to co-isolation and co-fragmentation of co-eluting peptides, which may necessitate additional correction strategies such as MS3 or SPS-MS3.
Use Case Comparison: When to Choose What
1. Exploratory Proteomics in Heterogeneous Clinical Samples → LFQ Preferred
In early-stage discovery studies involving heterogeneous samples such as patient-derived tissues, serum, or cerebrospinal fluid, label-free quantification offers key advantages. Since clinical samples are often limited in quantity and challenging to label, LFQ enables direct analysis without chemical modification. Moreover, it provides a broad dynamic range suitable for detecting both high- and low-abundance proteins, albeit with careful normalization and quality control.
Case: Identifying potential biomarkers in plasma samples from a small cohort of disease and control subjects.
The 2024 study used label-free quantification (LFQ) mass spectrometry to profile changes in plasma protein expression in obese patients before and after treatment with liraglutide. The study included 20 patients and compared pre- and post-treatment plasma samples, identifying 1,019 proteins in total, with 151 showing significant differential expression. These proteins were primarily involved in pathways related to lipid metabolism, inflammation, oxidative stress, and cytoskeletal organization. The findings suggest that liraglutide may exert its metabolic benefits through multi-pathway modulation of both inflammatory and metabolic proteins. This work demonstrates the strengths of LFQ in exploratory clinical proteomics, particularly when dealing with heterogeneous samples, small cohort sizes, and limited compatibility with labeling methods. Compared to techniques like TMT, LFQ offers greater flexibility and ease of use in early discovery studies, while still delivering reliable quantification through standardized normalization and quality control workflows.
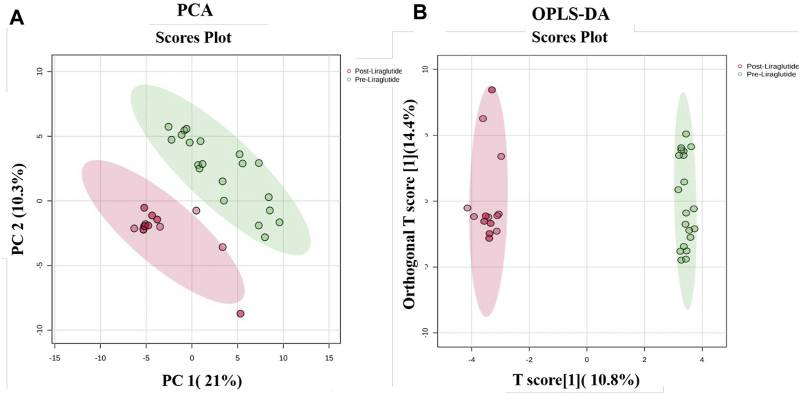
Figure4. Multivariate Analysis of Proteomic Profiles
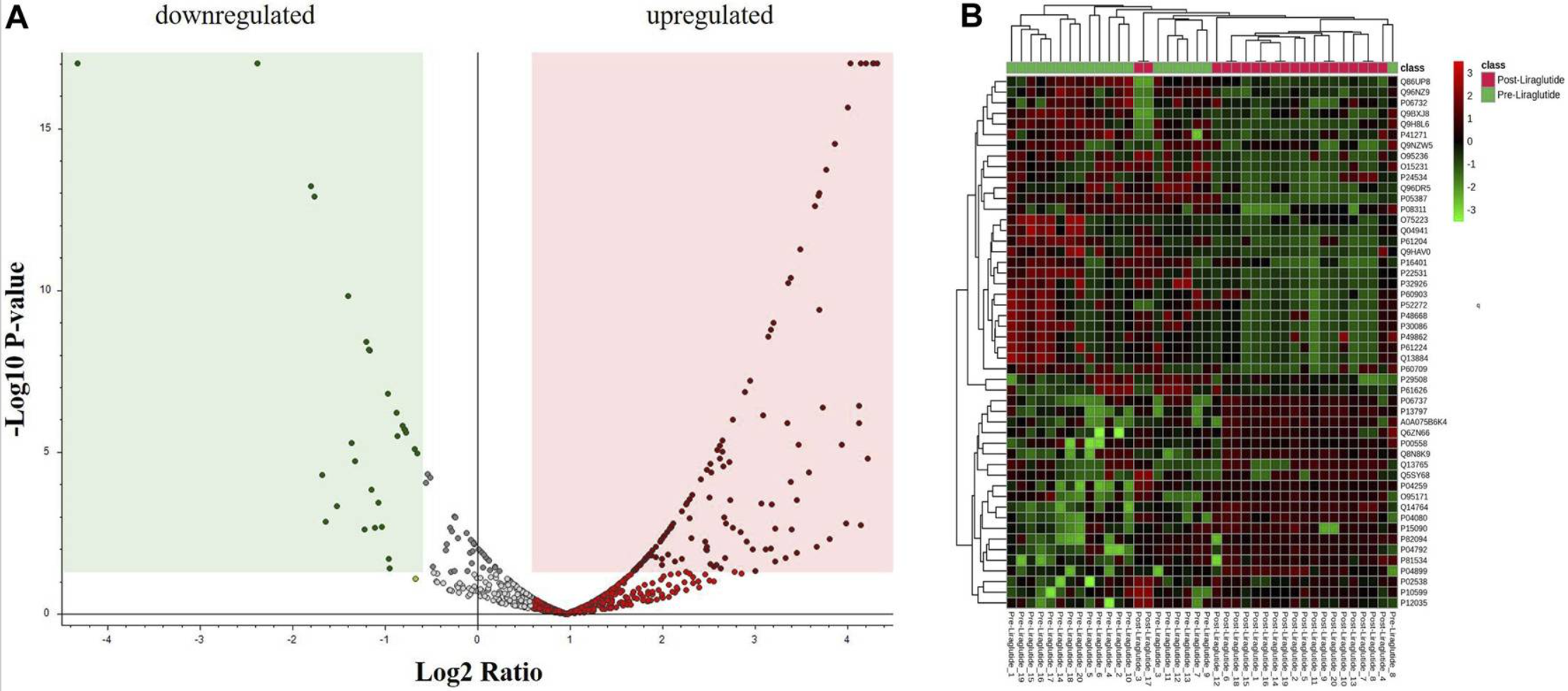
Figure5. Volcano Plot, Hierarchical Clustering and Heatmap Analysis of Identified Proteins
2. Large-Scale Comparative Studies with Multiple Conditions → TMT/iTRAQ Recommended
When analyzing multiple experimental conditions or biological replicates in a single study—such as drug screening, dose–response testing, or time-course profiling—isobaric labeling is the preferred choice. Techniques like TMT allow for the simultaneous analysis of up to 18 samples in a single run, minimizing batch effects and ensuring consistent quantification across conditions.
Case: Expanding the landscape of aging via orbitrap astral mass spectrometry and tandem mass tag integration
In a 2025 study published in Nature Communications, researchers leveraged the high-throughput capabilities of Orbitrap Astral mass spectrometry combined with TMT 18-plex labeling to perform a comprehensive proteomic profiling of multiple mouse tissues across aging stages. By simultaneously analyzing up to 18 samples in a single LC-MS/MS run, the study minimized batch effects and achieved consistent, high-depth quantification of over 12,000 proteins. The results revealed both linear and non-linear age-associated proteomic changes, with distinct patterns observed across brain regions and the kidney, including sex-specific differences. This work exemplifies how multiplexed quantitative proteomics enables systematic comparison across multiple biological conditions, offering valuable insights into tissue-specific aging mechanisms while establishing a scalable workflow suitable for large-cohort, condition-rich studies such as drug screening and time-course analyses.
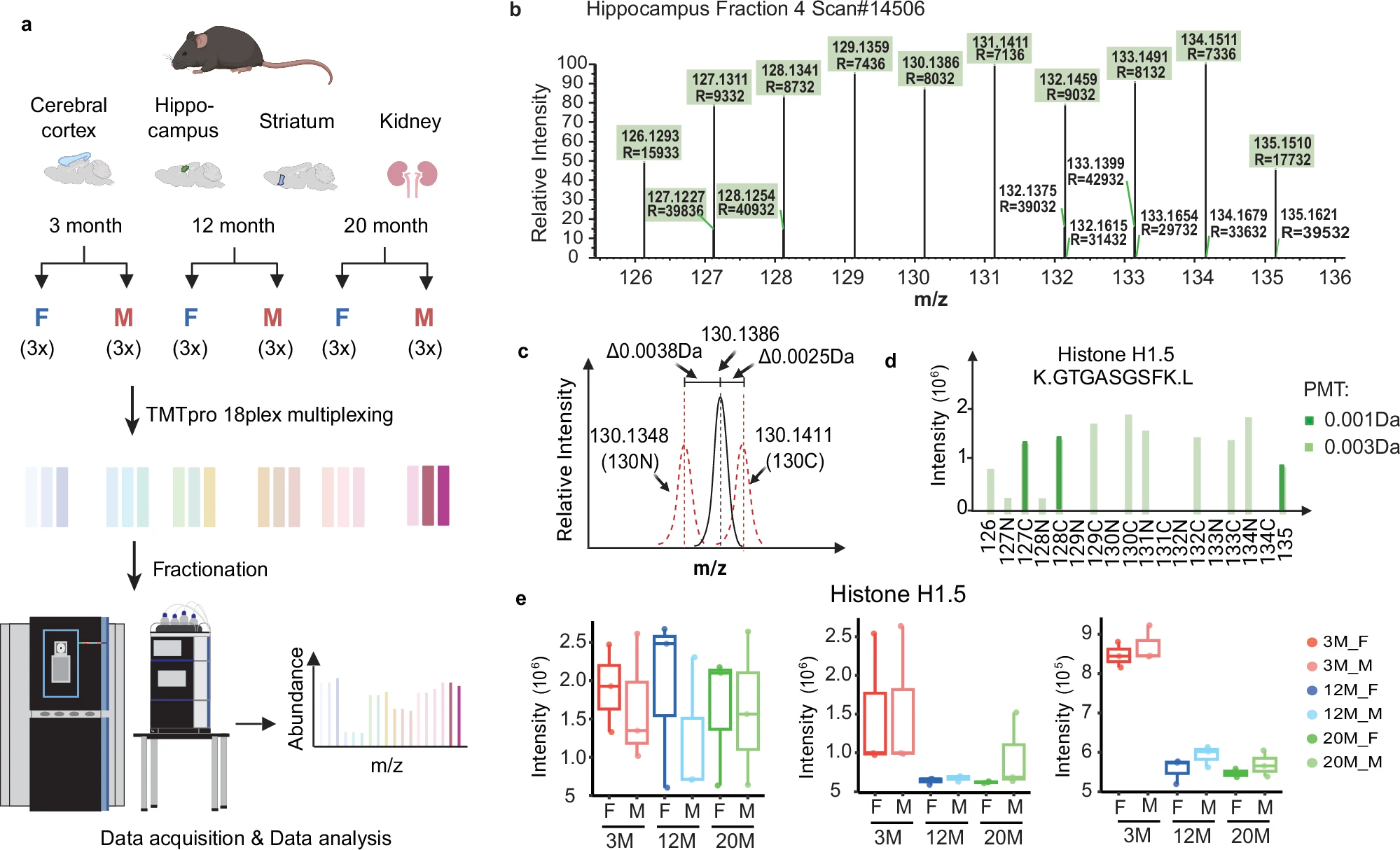
Keele, G.R. et al. Nat Commun. 2025.
Figure 6. Peptide Filtering Enhances TMT Quantification Accuracy with Orbitrap Astral Mass Spectrometry
How to order?







