Collisional Cross-Section Measurement Service
MtoZ Biolabs provides a professional Collisional Cross-Section Measurement Service to help researchers and industry clients obtain precise structural information on biomolecules and small molecules. By combining ion mobility spectrometry with high-resolution mass spectrometry, we deliver reliable collisional cross-section (CCS) values that complement mass and fragmentation data, enabling deeper insights into molecular conformation and dynamics.
At the microscopic level, molecular collisions play a critical role in determining how particles move and interact. When ions travel through a neutral gas under an electric field, they undergo repeated collisions with gas molecules. The frequency and intensity of these collisions depend on the ion's size, shape, and charge. This behavior provides a unique way to probe molecular architecture beyond mass-to-charge information. The physical parameter used to describe this interaction is known as the collisional cross-section (CCS), which reflects the effective surface area of an ion as it drifts through the gas medium. A larger CCS means more collisions and slower movement through the gas.
Unlike conventional mass spectrometry, which primarily measures mass, CCS adds an orthogonal dimension of data by capturing molecular size and shape. This allows researchers to distinguish structural isomers, validate computational models, and investigate conformational changes that influence biological activity, interactions, and stability.
Services at MtoZ Biolabs
MtoZ Biolabs' Collisional Cross-Section Measurement Service includes the use of ion mobility-based strategies to characterize molecular cross-sections and conformational changes, supported by hydrogen–deuterium exchange approaches for probing higher-order structures. In addition, we apply time-resolved ion mobility methods to capture dynamic conformational transitions, and we employ high-resolution mass spectrometry to refine CCS determinations for enhanced accuracy. Through these approaches, MtoZ Biolabs can distinguish structural isomers, investigate protein folding and unfolding, explore metabolite structures, and monitor biomolecular interactions.
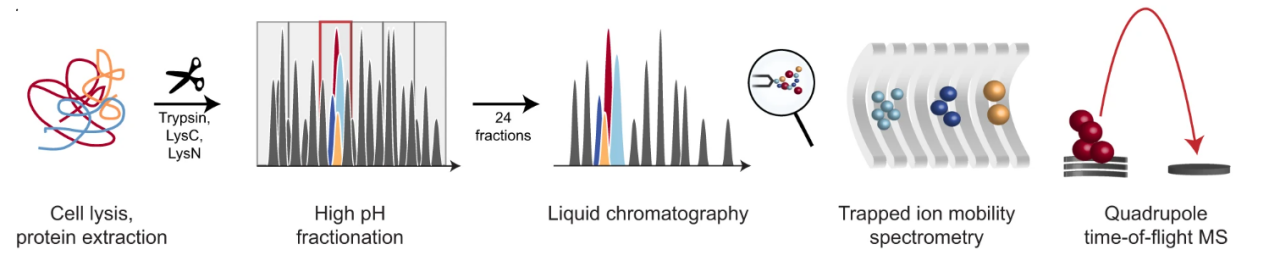
Meier, F. et al. Nat Commun. 2021.
Figure 1. Workflow for Large-Scale Peptide Collisional Cross-Section Measurement
Service Advantages
✔️Extensive expertise in ion mobility mass spectrometry and CCS measurement
✔️Proven track record across metabolomics, lipidomics, and pharmaceuticals
✔️Customized workflows tailored to client-specific goals and sample types
✔️Rigorous quality control for reproducible and publication-ready results
✔️End-to-end support from sample preparation consultation to data interpretation
Sample Submission Suggestions
1. Sample Types
Tissues, biofluids such as serum, plasma, urine, cerebrospinal fluid, cultured cells or lysates, microbial cultures, plant tissues, and purified extracts or fractions.
2. Recommended Amounts
The required amount depends on target analytes and matrix complexity. Typically, at least 200 μL of liquid sample or 200 mg of solid sample is required. Final quantities will be confirmed during project consultation.
3. Sample Condition
Samples should be free of detergents, salts, and other contaminants that may interfere with ionization.
4. Storage and Transport
Biological samples should be frozen at –80°C and shipped on dry ice. Minimize freeze–thaw cycles.
5. Documentation
Please provide detailed information on sample composition, concentration, and any prior analytical data to facilitate optimal analysis.
Clients are encouraged to contact MtoZ Biolabs for customized sample preparation advice.
FAQs
Q1: Does the Ion Charge State Affect CCS Measurement?
Yes, ions with different charge states can adopt distinct conformations, leading to variations in CCS values. This property is useful for studying protein unfolding and charge-dependent structures.
Q2: Can This Service Support Metabolite Identification in Untargeted Studies?
Yes, CCS is particularly valuable in untargeted metabolomics, where it helps separate and identify unknown metabolites that may overlap in m/z.
How to order?







