Co‑immunoprecipitation (Co‑IP) Overview
-
Dependence on antibody specificity and quality: Non-specific binding may result in false-positive signals, and antibody performance directly influences experimental reliability.
-
Background interference: Non-specific adsorption of proteins in lysates can compromise signal clarity.
-
Limited sensitivity to weak or transient interactions: Compared with cross-linking approaches or mass spectrometry-based strategies, Co-IP is less effective in capturing low-affinity complexes.
-
Utilization of high-affinity, rigorously validated antibodies.
-
Application of chemical cross-linkers (e.g., DSS, DSP) to stabilize weak interactions.
-
Employment of magnetic bead-based systems to enhance washing efficiency and reduce background noise.
-
Integration with mass spectrometry to improve interaction coverage and identification depth.
Protein-protein interactions form the molecular basis of cellular signal transduction, metabolic regulation, and disease pathogenesis. As a classical approach for investigating endogenous protein interactions, co-immunoprecipitation (Co-IP) remains widely applied in molecular biology and proteomics research owing to its operational robustness, high specificity, and broad applicability. In particular, Co-IP is well suited for validating interactions between defined protein pairs.
What Is Co-immunoprecipitation (Co-IP)?
Co-immunoprecipitation (Co-IP) is a well-established biochemical technique used to investigate protein-protein interactions (PPIs). This method relies on the specific binding between antigens and antibodies. By immunoprecipitating a designated “bait” protein, interacting “prey” proteins associated with the bait are co-enriched, thereby providing indirect evidence for physiologically relevant protein interactions.
Co-IP is among the most widely employed techniques for PPI analysis and is particularly advantageous for validating protein interactions under native conditions. Compared with in vitro systems, such as yeast two-hybrid assays, Co-IP more accurately reflects the composition of protein complexes under endogenous or near-physiological environments.
Core Principles of Co-IP
The standard Co-IP workflow consists of the following key steps:
1. Cell Lysis: Cells or tissues of interest are lysed in appropriate buffers under mild conditions to preserve the native conformation of protein complexes and their interaction states.
2. Antibody Incubation: A target-specific antibody is introduced to bind the protein of interest, forming an antibody-protein complex.
3. Capture by Solid Support: Protein A/G magnetic beads, which bind to the Fc region of antibodies, are used to immobilize and isolate the complex.
4. Washing and Elution: Non-specific binders are removed through washing steps, while interacting proteins are retained and subsequently eluted.
5. Downstream Analysis: Interacting proteins are commonly detected by Western blotting, and mass spectrometry can be employed to further characterize complex composition.
Advantages of Co-IP
1. Physiological Relevance: Mild lysis conditions facilitate the preservation of native protein complexes.
2. Technological Maturity: The workflow is well established and readily implemented in most molecular biology laboratories.
3. Compatibility with Diverse Downstream Analyses: Co-IP can be integrated with Western blotting, mass spectrometry, enzyme activity assays, and other analytical techniques for qualitative and quantitative evaluation of protein interactions.
Technical Limitations and Optimization Strategies
Despite its widespread use, Co-IP presents certain limitations, particularly in high-throughput applications and in the detection of weak or transient protein interactions.
1. Major Technical Challenges
2. Optimization Strategies
Comparison of Co-IP with Other Protein Interaction Techniques
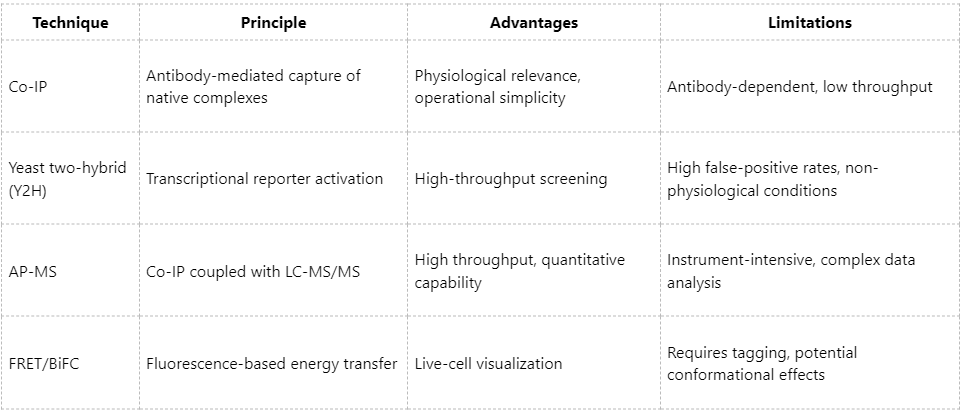
Overall, Co-IP is best suited for validating candidate protein interactions in hypothesis-driven studies involving known targets. For exploratory analyses or large-scale interactome mapping, affinity purification-mass spectrometry (AP-MS) and related network-based approaches offer distinct advantages.
Co-IP Coupled with Mass Spectrometry: Advancing Interaction Studies
The integration of Co-IP with mass spectrometry (Co-IP-MS) has emerged as a powerful strategy for comprehensive characterization of protein complexes. The workflow typically includes:
1. Immuno-enrichment of protein complexes using target-specific antibodies.
2. Proteolytic digestion following washing and elution.
3. Identification and quantification of interacting proteins by LC-MS/MS.
4. Analysis of interaction dynamics using quantitative approaches such as TMT labeling or label-free methods.
Co-IP-MS enables not only the confirmation of known interactions but also the discovery of previously uncharacterized interaction networks, with broad applications in signaling pathway analysis, disease mechanism studies, and drug target discovery.
Co-IP and Interactomics Services at MtoZ Biolabs
Building on extensive experience in immuno-enrichment and mass spectrometry-based proteomics, MtoZ Biolabs has established an integrated Co-IP + LC-MS/MS analytical platform applicable to a wide range of research scenarios, including:
1. Characterization of protein interactions at critical signaling pathway nodes.
2. Comparative interactome analysis of disease-associated mutant proteins.
3. Quantitative assessment of interaction network alterations following pharmacological interventions.
4. Screening and functional validation of novel molecular targets.
By leveraging high-resolution Orbitrap mass spectrometry, optimized immuno-enrichment protocols, and robust data analysis pipelines, we provide researchers with reproducible, high-coverage, and high-specificity interactome datasets.
As a classical yet continually evolving technique, Co-IP remains indispensable in protein interaction research. With ongoing advances in mass spectrometry and computational methodologies, the scope of Co-IP applications continues to expand. Selecting appropriate interaction analysis strategies based on experimental objectives, sample characteristics, and available resources is critical. MtoZ Biolabs is committed to supporting life science research through rigorous technological platforms and professional analytical expertise, facilitating deeper insights into the molecular mechanisms underlying cellular regulation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







