Carbonylation Proteomics Service
Carbonylation proteomics analysis is a proteomic technique dedicated to detecting and characterizing protein carbonylation modifications. Protein carbonylation is an irreversible oxidative modification primarily mediated by reactive oxygen species (ROS), commonly occurring on lysine, proline, arginine, and threonine residues, resulting in the formation of aldehyde or ketone groups. This modification can be identified by high-resolution mass spectrometry combined with advanced techniques such as ELISA and Western blot (WB) to determine carbonylation sites and variation levels. Bioinformatics analysis further reveals its functional impact on cellular homeostasis imbalance.
Carbonylation proteomics service is widely applied in studies related to oxidative stress-associated diseases, such as neurodegenerative disorders, cardiovascular diseases, diabetes, inflammatory conditions, and aging mechanisms. This service enables the construction of protein oxidative damage profiles, identification of potential biomarkers, evaluation of antioxidant drug efficacy, and supports the elucidation of cellular injury mechanisms and stress response regulatory networks.
Cabiscol, E. et al. Mass Spectrometry Reviews, 2013.
Figure 1. Physiological Function of Carbonylated Proteins.
Services at MtoZ Biolabs
Based on a high-resolution mass spectrometry platform and multiple protein carbonylation detection techniques, the carbonylation proteomics service launched by MtoZ Biolabs systematically analyzes protein carbonyl modifications induced by oxidative stress. This service combines chemical derivatization with mass spectrometry to precisely identify modification sites and peptide sequences, and is further supported by validation and enrichment strategies such as Western blotting, ELISA, and chemical probe-based enrichment, thereby enhancing the detection of low-abundance modified proteins. The resulting data include carbonylation sites, relative abundance, spectral information, and functional annotations, making this service applicable to oxidative stress mechanism research, biomarker discovery, and drug efficacy evaluation. The analytical methods include:
1. Mass Spectrometry (MS)
Carbonyl modifications on proteins are derivatized into detectable forms, followed by high-resolution LC-MS/MS for peptide identification and modification site localization. This method offers high sensitivity and broad coverage, enabling both qualitative and quantitative analysis, and is considered the core technology in carbonylation proteomics research.
2. Western Blot
Post-derivatization, DNP-specific antibodies are used to detect carbonylated proteins. This method is simple and produces visually interpretable results, making it suitable for targeted validation, but it does not provide site-specific information or accurate quantification.
3. ELISA
Enzyme-linked immunosorbent assays using anti-carbonyl antibodies are employed to assess the total carbonylation levels in samples. While suitable for high-throughput screening or preliminary assessment, this method lacks resolution for identifying specific proteins or modification sites.
4. 2D Gel Electrophoresis with DNPH Staining
Proteins are separated by two-dimensional gel electrophoresis and visualized via DNPH staining or Western blotting, revealing global carbonylation profiles. This technique is suitable for analyzing large-scale expression changes but has limited sensitivity and is not ideal for detecting low-abundance proteins or providing precise quantification.
5. Chemical Probe Labeling
Carbonylated proteins are selectively labeled using chemical probes, enriched via affinity capture, and analyzed by mass spectrometry. This method significantly improves detection of low-abundance carbonylated proteins and is particularly useful for enrichment and deep profiling in complex biological matrices.
Analysis Workflow
1. Protein Extraction and Digestion
Target proteins are extracted from cells, tissues, or purified protein samples, followed by reduction and alkylation. Trypsin digestion is then performed to generate peptides suitable for mass spectrometry detection while preserving carbonylation modifications.
2. Carbonylation Derivatization and Enrichment
Carbonyl modifications are derivatized using chemical derivatization to form hydrazone derivatives. Carbonylated peptides are then selectively enriched using antibody-based or solid-phase extraction methods.
3. Multi-Platform Detection and Analysis
A combination of LC-MS/MS, ELISA, and Western Blot techniques is used to accurately identify carbonylation modification sites and quantify their abundance, improving detection accuracy and data coverage.
4. Data Processing and Result Output
Specialized databases and analytical software are employed to identify and quantify modification sites. The final report includes modification positions, spectral data, relative abundance, and functional annotation.
Sample Submission Suggestions
1. Sample Types
Supports a variety of sample forms including cells, tissues, plasma/serum, and purified proteins. Oxidative stress-related models are recommended to enhance the detection efficiency of carbonylation modifications.
2. Buffer Requirements
Avoid components such as SDS, glycerol, and EDTA that may interfere with mass spectrometry or derivatization reactions. MS-compatible, non-reducing buffer systems are recommended.
3. Sample Storage and Transport
Samples should be stored at –80°C and transported on dry ice. Avoid repeated freeze-thaw cycles to ensure the stability of carbonylation modifications and the reliability of analytical results.
Service Advantages
1. High Sensitivity Detection
Leveraging high-resolution mass spectrometry platforms combined with specific derivatization and enrichment techniques, our service accurately identifies low-abundance carbonylation modifications, enabling precise analysis of complex samples.
2. Multi-Strategy Enrichment
Employs various enrichment approaches, such as DNPH-based chemical derivatization and affinity capture, to improve the detection efficiency and coverage depth of modified peptides.
3. Data Visualization Output
Delivers comprehensive datasets including peptide sequences, modification sites, spectral images, and abundance distribution, along with graphical reports for easy interpretation and comparison.
4. One-Stop Technical Service
Covers the complete workflow from sample preparation, modification detection, and quantitative analysis to bioinformatics interpretation, supporting a wide range of research and drug development needs.
Applications
1. Oxidative Stress Mechanism Research
The carbonylation proteomics service can be used to monitor the occurrence and changes of protein carbonylation modifications under oxidative stress conditions, revealing their roles in cellular stress responses and damage pathways.
2. Aging and Neurodegenerative Disease Research
Carbonylation modifications are highly enriched in aging and in neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease. This service helps explore the association between carbonylation and protein dysfunction.
3. Inflammation and Chronic Disease Mechanism Analysis
By analyzing the distribution patterns of protein carbonylation in chronic diseases such as diabetes and atherosclerosis, this service supports the investigation of disease pathogenesis and the identification of potential therapeutic targets.
4. Environmental Toxicology and Exposure Response Assessment
The carbonylation proteomics service is suitable for evaluating carbonylation responses triggered by environmental pollutants (such as heavy metals and particulate matter), helping to uncover their impact on cellular oxidative damage.
Case Study
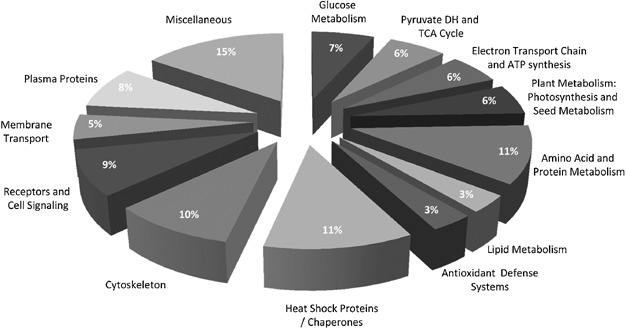
1. Proteomic and Carbonylation Profile Analysis at the Critical Node of Seed Ageing in Oryza sativa
This study aimed to elucidate the proteomic changes and carbonylation profiles at the critical node of seed aging in rice, in order to reveal the molecular mechanisms underlying seed vigor decline. Rice seeds stored for different durations were selected as the study subjects. Protein expression was analyzed using two-dimensional gel electrophoresis (2-DE) combined with MALDI-TOF/TOF mass spectrometry, while carbonylation levels were assessed via DNPH derivatization. A total of 56 differentially expressed proteins were identified, primarily associated with energy metabolism, antioxidative processes, defense responses, and protein synthesis. Many of these proteins exhibited significant carbonylation accumulation during aging. The findings suggest that protein carbonylation serves as a key marker of oxidative damage in seed aging and may impair the function of essential metabolic and defense proteins, ultimately leading to loss of vigor. This provides a theoretical foundation for germplasm conservation and seed longevity regulation in crops.
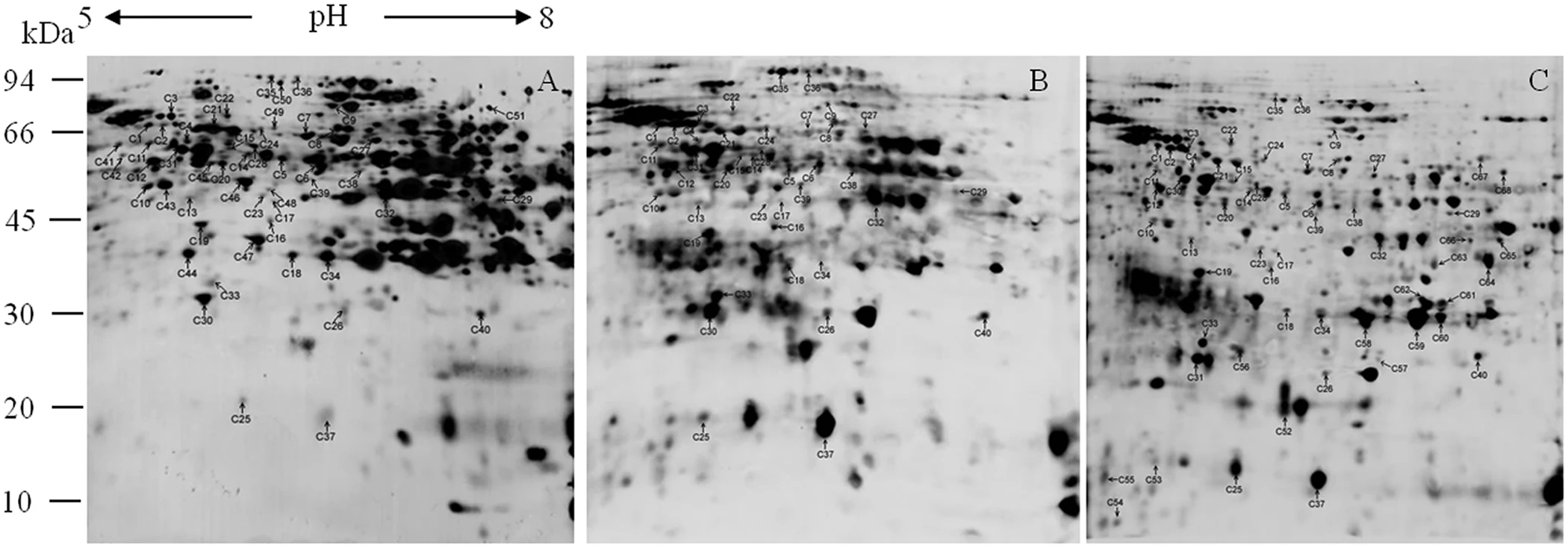
Yin, G K. et al. Scientific Reports, 2017.
Figure 2. Carbonylated Proteins from Rice Seeds in 2D Blots.
2. Proteomic Analysis and Protein Carbonylation Profile in Trained and Untrained Rat Muscles
This study aimed to evaluate the effects of aerobic training on protein expression and carbonylation modification in rat skeletal muscle, exploring its underlying antioxidant mechanisms. Trained and untrained rats were used as experimental subjects. Proteins from the quadriceps were extracted and analyzed using two-dimensional gel electrophoresis (2-DE) combined with MALDI-TOF/TOF mass spectrometry, while carbonylation levels were assessed using DNPH derivatization and immunodetection methods. The results showed that multiple proteins involved in energy metabolism, muscle contraction, and antioxidant stress response were upregulated in the trained group, and overall protein carbonylation levels were significantly reduced. The study concludes that aerobic training enhances the antioxidant capacity of skeletal muscle by modulating protein expression and reducing oxidative damage.
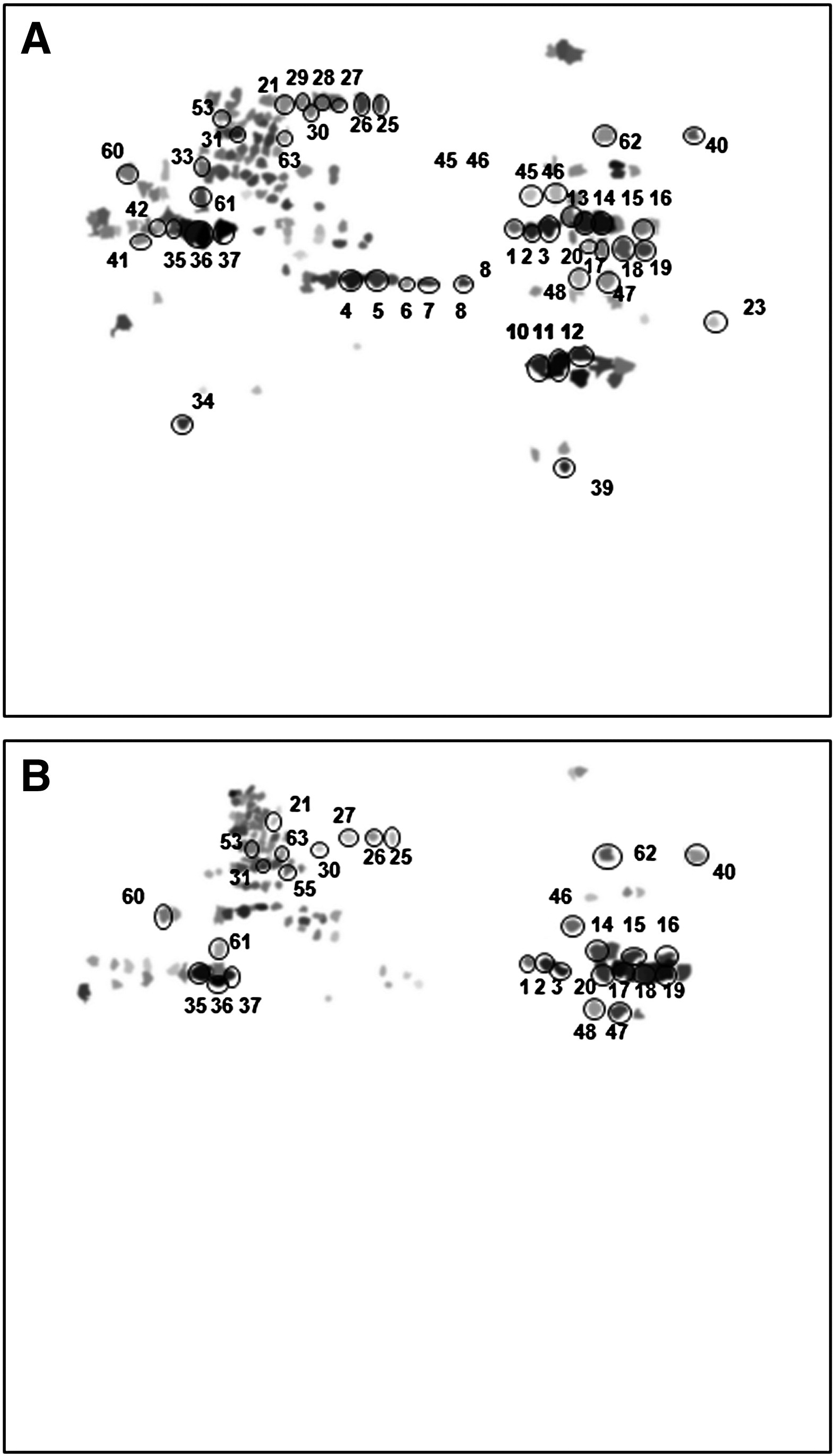
Magherini, F. et al. Journal of Proteomics, 2012.
Figure 3. Synthetic Gels Representing the Carbonylation Pattern of Soleus (Panel A) and Tibialis Anterior (Panel B) Muscles.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
FAQ
Q1: Is Low-Abundance Carbonylated Protein Detection Supported?
A1: Yes. By combining specific derivatization methods (such as DNPH labeling), affinity enrichment strategies, and high-resolution mass spectrometry platforms, we can effectively identify low-abundance carbonylation modifications.
Q2: Is Quantitative Analysis Supported?
A2: Yes. Depending on the research needs, we offer various quantification strategies such as Label-Free, TMT, or iTRAQ to compare carbonylation modification levels across different sample groups.
How to order?







