A Comprehensive Overview of Tandem Mass Tag (TMT)-Based Quantitative Proteomics
-
Multiplexing capability: Supports simultaneous analysis of 10, 11, 16, or 18 samples.
-
High throughput with minimal batch effects, reducing systematic bias across experimental groups.
-
Excellent quantification consistency, well-suited for complex designs such as clinical cohorts, time-course studies, and treatment-control comparisons.
-
Reactive group: Covalently binds to the peptide’s N-terminus or lysine side chains.
-
Mass normalizer: Balances the overall mass of labeled peptides to ensure they remain isobaric at the MS1 level.
-
Reporter ion: Released upon fragmentation during MS2 or MS3, enabling quantification.
-
Differential protein expression between tumor tissues and adjacent normal tissues;
-
Comparative plasma proteomic profiling in autoimmune disease patients.
-
Extensive proteome coverage: Quantification of >7,000 proteins and >50,000 peptides on average
-
High quantification accuracy: Incorporation of MS3-based correction algorithms to minimize isotopic interference
-
High data completeness: Multi-channel sample integration reduces the incidence of missing values
-
Support for large cohorts: Compatibility with TMTpro 16/18plex formats for high-throughput projects
-
Customized data analysis: End-to-end services including functional annotation, pathway enrichment, and in-depth bioinformatics interpretation
Accurate quantification is fundamental to uncovering biological differences in proteomics research. As experimental designs involving multiple samples and time points become increasingly prevalent, traditional label-free approaches have demonstrated limitations in terms of consistency and throughput. Tandem Mass Tag (TMT)-based quantitative proteomics has emerged as a powerful alternative, offering high-throughput capabilities, low variability, and parallel sample analysis. These features make it an indispensable tool for multi-omics studies and biomarker discovery in life sciences.
What is TMT-Based Quantitative Proteomics?
TMT (Tandem Mass Tag) is an isobaric labeling technique based on mass spectrometry, enabling the relative quantification of up to 16 or more samples simultaneously. Developed by Thermo Fisher Scientific, the method has evolved into the TMTpro™ 18plex version, facilitating parallel quantification of up to 18 samples, significantly improving the efficiency of large-scale studies.
Key advantages of TMT-based quantification include:
Principles of TMT Technology
1. Structure of TMT Reagents
Each TMT reagent is composed of three functional components:
All TMT tags are isobaric, having the same total mass, and thus indistinguishable in MS1 scans. However, upon fragmentation, they release reporter ions with distinct masses, which can be detected at the MS2 or MS3 level for accurate relative quantification.
2. Overview of Experimental Workflow
A typical TMT-based proteomics experiment comprises the following key steps:
(1) Protein Extraction and Digestion
Proteins are extracted from each biological sample and enzymatically digested, commonly using trypsin, to generate peptides.
(2) TMT Labeling
Peptides derived from each sample are labeled with a distinct TMT reagent.
(3) Sample Pooling and Peptide Fractionation
Labeled samples are mixed in equal amounts. Fractionation is performed using high-pH reverse-phase liquid chromatography (High-pH RPLC) or strong cation exchange (SCX) chromatography to enhance peptide coverage.
(4) LC-MS/MS Analysis
Tandem mass spectrometry is performed using high-resolution platforms such as the Orbitrap Fusion Lumos or Exploris 480. MS3 acquisition mode is commonly employed to minimize co-isolation interference and improve quantification precision.
(5) Data Processing
Software tools such as Proteome Discoverer are used for peptide identification, extraction of reporter ion intensities, and subsequent relative quantification.
Technical Comparison: TMT vs. Label-Free Quantification
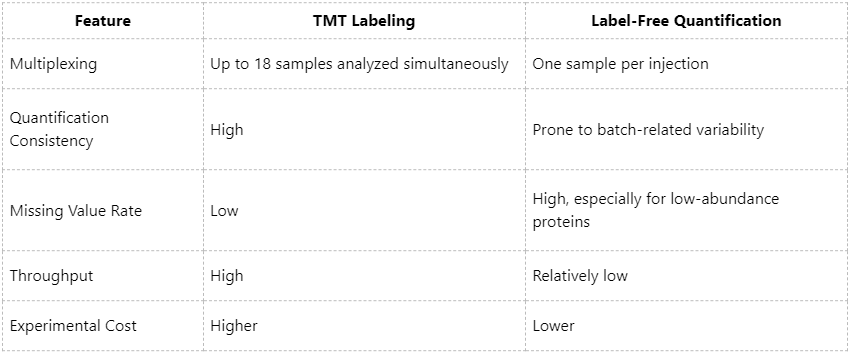
Given these characteristics, TMT-based quantification is particularly advantageous for studies requiring accurate inter-sample comparison, such as disease subtyping, drug response evaluation, and time-resolved profiling.
Application Scenarios: Broad Utility of TMT in Life Sciences
1. Disease Mechanism Elucidation
TMT-based quantification enables precise identification of differentially expressed proteins between disease and control groups, contributing to the understanding of pathological mechanisms.
Examples include:
2. Drug Screening and Target Validation
TMT can assess proteomic changes before and after drug treatment, aiding in elucidating drug mechanisms and validating therapeutic targets.
3. Biomarker Discovery
Large-scale TMT analyses can reveal candidate protein biomarkers with reproducible expression alterations, supporting downstream clinical translation.
4. Time-Course Studies
The TMT platform is well-suited for time-series experiments (e.g., 0h, 6h, 12h, 24h), enabling dynamic profiling of protein expression changes.
TMT Quantitative Proteomics Solutions at MtoZ Biolabs
At MtoZ Biolabs, we integrate cutting-edge mass spectrometry platforms (e.g., Orbitrap Exploris 480, Fusion Lumos) with an optimized TMT data processing pipeline to deliver comprehensive services to both academic and industrial clients:
Whether your goal is to advance basic biological research, discover biomarkers, or validate drug targets, MtoZ Biolabs offers robust and scalable solutions for TMT-based proteomic studies.
With the ongoing advancement of life sciences, the demand for precise characterization of protein expression dynamics continues to rise. TMT-based quantitative proteomics has become a cornerstone of modern proteomic research, owing to its multiplexing capacity, throughput efficiency, and quantification accuracy. At MtoZ Biolabs, we emphasize not only the generation of high-quality mass spectrometry data but also the extraction of meaningful biological insights. Through rigorous experimental workflows and comprehensive bioinformatics support, we aim to empower researchers in making impactful discoveries in the field of proteomics.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







