What Is Shotgun Protein Identification and How Does It Differ from Traditional Methods?
- Oncology: Comparative proteomic profiling of cancerous and normal tissues to elucidate oncogenic mechanisms.
- Neuroscience: Proteomic analysis of brain tissue and cerebrospinal fluid for early diagnosis of neurodegenerative diseases such as Alzheimer’s disease.
- Infection and Immunology: Characterization of host–pathogen interaction mechanisms.
- Microbiomics: Functional proteome analysis of microbial communities from environments such as the gut and oral cavity.
- Drug Discovery and Toxicology: Assessment of proteome-level responses to pharmacological interventions, aiding in target identification and mechanistic interpretation.
- Advanced high-resolution mass spectrometry platforms, including Orbitrap Exploris 480, Q Exactive HF-X, and timsTOF Pro 2.
- A proprietary enzymatic digestion system and optimized LC–MS configurations, enhancing both peptide identification rates and data reproducibility.
- Support for multiple quantification methods (Label-Free, TMT, DIA) to accommodate diverse experimental designs.
- Comprehensive bioinformatics services, including GO/KEGG pathway enrichment, PPI network construction, and disease association analysis.
- Final reports meeting SCI-indexed publication standards, facilitating use in manuscript submission and grant applications.
Shotgun protein identification is a large-scale protein identification strategy that involves enzymatically digesting complex protein mixtures (commonly using trypsin) into peptides, followed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. The resulting spectra are then searched against protein databases to infer the identities of the original proteins. The term shotgun refers to its non-targeted analytical approach; rather than analyzing proteins one by one, it comprehensively identifies all proteins present in a sample. This strategy replaces traditional single-protein analytical methods with a holistic workflow of global enzymatic digestion combined with large-scale peptide analysis, enabling simultaneous identification of thousands of proteins. Such an approach provides a robust technical foundation for biomarker discovery, drug mechanism elucidation, and integrative multi-omics research.
Core Workflow of Shotgun Protein Identification
1. Sample Preparation
Biological materials such as cells, tissues, or body fluids are lysed, and contaminants (e.g., salts, lipids) are removed before proteins are extracted for further processing.
2. Protein Digestion
Proteins are typically digested with trypsin to generate smaller peptides suitable for mass spectrometry analysis.
3. Peptide Separation (LC)
High-performance liquid chromatography (HPLC) is employed to separate complex peptide mixtures, thereby reducing the analytical burden on the mass spectrometer at any given moment.
4. Mass Spectrometry Analysis (MS/MS)
Tandem mass spectrometry determines the mass-to-charge ratios (m/z) of peptides. Fragment ion spectra are then interpreted to deduce the amino acid sequences of the peptides.
5. Database Search and Protein Identification
Software tools such as SEQUEST, Mascot, and MaxQuant compare experimental spectra against reference protein databases (e.g., UniProt) to achieve protein identification and quantification.
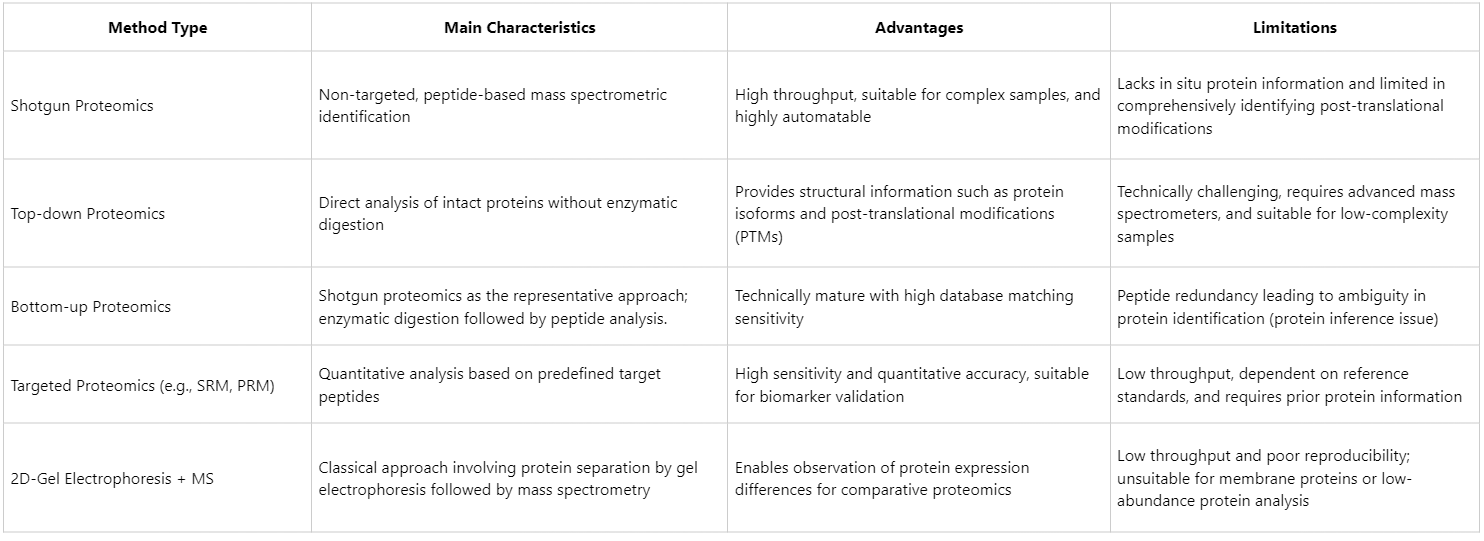
Comparison Between Shotgun Protein Identification and Other Analytical Methods
Core Advantages of Shotgun Proteomics
1. High-Throughput Coverage
Shotgun proteomics enables the simultaneous detection of thousands of proteins, making it highly suitable for complex biological samples such as tumor tissues, serum, cerebrospinal fluid, and microbial communities.
2. Automation and Reproducibility
By integrating advanced automated sample-preparation systems with stable mass spectrometry platforms, the workflow ensures strong batch-to-batch reproducibility, facilitating large-scale and multi-center studies.
3. Compatibility with Multi-Omics Platforms
The shotgun approach complements transcriptomic, metabolomic, and epigenomic datasets, serving as a key integrative node in systems biology.
4. Flexible Quantification Strategies
It supports both labeling-based quantification (e.g., TMT, iTRAQ) and label-free quantification (LFQ), allowing researchers to tailor experiments to specific study designs.
Application Scenarios of Shotgun Protein Identification: From Basic Research to Clinical Translation
MtoZ Biolabs Enabling High-Quality Proteomics Research
MtoZ Biolabs has developed a comprehensive end-to-end solution for shotgun protein identification, encompassing sample processing, peptide optimization, mass spectrometry detection, and data analysis.
Key technical highlights include:
Through continuous optimization of experimental workflows and data-analysis algorithms, the company achieves identification of over 10,000 proteins per sample, with quantitative protein CV values below 15%, ensuring high coverage and stability of proteomic datasets.
With continuous advancements in mass spectrometry technologies and computational algorithms, the shotgun protein identification strategy is expected to evolve further, enhancing both the depth and accuracy of proteome profiling. Collaborative efforts with MtoZ Biolabs can facilitate deeper exploration of proteomic landscapes and contribute to advancing the frontiers of life science research.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







