What Is DIA Proteomics? A Complete Guide to Core Techniques
-
Data can be re-analyzed using updated algorithms or newly developed databases;
-
Suitable for establishing long-term spectral library assets of the proteome;
-
Particularly valuable in studies where sample re-collection is infeasible, such as clinical or animal research.
-
Library-based analysis: Coupled with DDA-generated project-specific spectral libraries, this approach offers high-accuracy identification;
-
Library-free analysis: Enables efficient data processing without the need for a pre-built library;
-
Hybrid strategies: Provide a balance between analytical depth and sample throughput.
-
Support for multiple instrument platforms (Orbitrap Exploris, TripleTOF 6600, timsTOF Pro)
-
A professional approach to spectral library construction, compatible with a variety of species including human, mouse, and plant
-
Value-added services including biological enrichment analysis, protein-protein interaction network construction, and target prediction
-
Provision of reusable data packages to facilitate supplementary analyses for future publications
As proteomics progresses toward high-throughput, high-precision, and large-scale cohort studies, the Data-Dependent Acquisition (DDA) strategy has increasingly revealed its limitations—namely, constrained scanning efficiency, limited reproducibility in quantification, and frequent omission of low-abundance proteins. In contrast, Data-Independent Acquisition (DIA) is emerging as a powerful approach for investigating complex biological systems. Compared to DDA, DIA not only improves quantification accuracy but also markedly enhances reproducibility and proteome coverage.
What is DIA proteomics? Why is it gaining prominence as the new mainstream in mass spectrometry? And what are the core technologies that underpin DIA-based analysis? This article aims to provide a comprehensive overview.
What Is DIA Proteomics? (Principle and Acquisition Strategy)
Data-Independent Acquisition (DIA) is an alternative mass spectrometry acquisition strategy. Its fundamental principle involves partitioning the full mass range into multiple predefined windows and fragmenting all ions within each window in a systematic manner. Unlike DDA, which selectively targets precursor ions—typically those of higher abundance—DIA collects fragmentation data across all ions in an unbiased fashion, thereby enabling more consistent and comprehensive proteomic profiling.
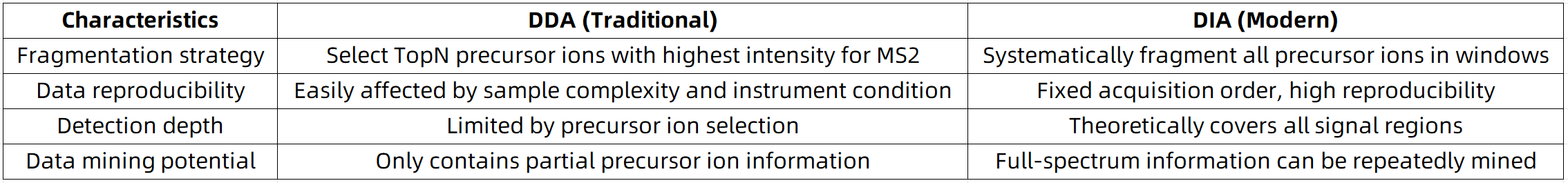
Table 1. Workflow Comparison: DDA vs. DIA
By retaining all potential ion information, DIA ensures both data completeness and reproducibility. This feature makes it especially well-suited for high-precision applications, including large-cohort studies, longitudinal analyses, and biomarker discovery.
Evolution and Mainstream Platforms of DIA Technology
Although DIA is not a new concept—its theoretical framework dates back to the 1990s—it was only in the past decade that the method became practically viable. This progress was driven by major advancements in mass spectrometry hardware and significant improvements in data analysis algorithms, enabling DIA to be applied with both high efficiency and precision.
1. Key Development Milestones
(1) 2012 – Introduction of SWATH-MS (SCIEX): Marked the first implementation of a window-based data-independent acquisition strategy, enabling systematic fragmentation of all precursor ions.
(2) 2014 – Launch of OpenSWATH: Pioneered the era of open-source solutions for DIA data analysis, broadening accessibility and reproducibility.
(3) 2018 – Release of Spectronaut and DIA-NN: Signified a turning point in the computational maturity of DIA, with algorithms reaching levels of robustness suitable for high-throughput applications.
(4) 2020 – Emergence of diaPASEF (Bruker): Integrated ion mobility separation with DIA acquisition, significantly enhancing both sensitivity and acquisition speed.
(5) 2021 – Adoption of Library-Free DIA Approaches: Enabled accurate, high-resolution quantification without the need for prior spectral libraries, facilitating broader and more flexible applications.
2. Mainstream DIA Mass Spectrometry Platforms

Table 2. Mainstream DIA Mass Spectrometry Platforms
MtoZ Biolabs offers a comprehensive, multi-platform DIA mass spectrometry infrastructure, including SWATH, diaPASEF, and SureQuant, supporting an end-to-end proteomics workflow. This includes DDA-based spectral library construction, DIA-based quantitative acquisition, and PRM-based validation.
Core Advantages of DIA Proteomics
1. Higher Detection Coverage
DIA avoids the selection bias inherent in DDA that tends to overlook low-abundance peptides. This makes it particularly well-suited for complex biological samples—such as tissues, plasma, and FFPE—where regulatory proteins and cytokines are often challenging to detect consistently.
2. Improved Data Reproducibility
With DIA’s fixed acquisition scheme, each sample yields comparable ion profiles, which facilitates robust reproducibility. This is especially advantageous for large-scale cohort studies or time-series experiments, ensuring data consistency across different batches, time points, and analytical instruments.
3. Greater Data Utilization Efficiency
DIA retains comprehensive MS/MS fragment data, enabling high potential for retrospective analysis:
4. Flexible Analytical Strategies (Library-Based / Library-Free)
DIA supports multiple analysis modes:
DIA Analysis Workflow: A Systematic Explanation from Acquisition to Quantification
1. Sample Preprocessing
A standard proteomics workflow—comprising lysis, enzymatic digestion, and purification—should be followed, with an emphasis on maintaining inter-batch consistency. Sample types compatible with DIA include cells, tissues, serum/plasma, and FFPE specimens.
2. DIA Mass Spectrometry Acquisition
An optimized windowing strategy—either fixed-width or variable-width—is applied, typically dividing the m/z range into 30 to 60 acquisition windows, depending on the mass spectrometry platform. Precursor ions are fragmented across the full mass range to generate comprehensive MS/MS data.
3. Data Processing Workflow
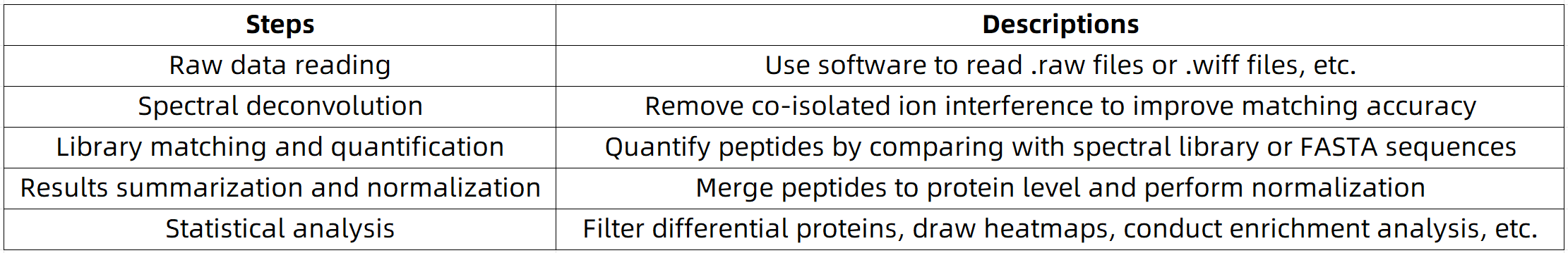
Table 3. DIA Data Processing Workflow
4. Commonly Used Analysis Software
(1) Spectronaut
(2) DIA-NN (free and open-source; computationally efficient and highly accurate)
(3) OpenSWATH (an early, widely adopted open-source tool)
(4) Skyline (well-suited for validation workflows; supports hybrid PRM/DIA analysis)
MtoZ Biolabs offers customized DIA data analysis services, including differential protein expression analysis, GO and KEGG pathway annotation, clustering, and protein–protein interaction network construction. The results can be delivered with high-quality figures suitable for publication in SCI-indexed journals.
MtoZ Biolabs: A High-Quality DIA Proteomics Service Platform
We have established an integrated DIA proteomics workflow encompassing the entire process from sample preparation to data mining, offering the following advantages:
The rapid advancement of DIA technology is accelerating the evolution of proteomics from an exploratory field to a quantitative science. This shift not only enhances data quality but also opens up new possibilities for systems biology and translational clinical research. Researchers are encouraged to contact us for complimentary support in DIA experimental design or to request example data packages to initiate high-throughput proteomic investigations.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







