What Is CUT&Tag? Principles, Workflow, and Advantages Explained
In recent years, the exploration of chromatin states has transitioned from ChIP-seq to more sensitive and efficient approaches. Among these, Cleavage Under Targets and Tagmentation (CUT&Tag) has gained significant prominence due to its high sensitivity, low background noise, and compatibility with high-throughput applications, making it a preferred technique in the field of chromatin epigenetics. This review provides a comprehensive examination of the principles, experimental workflow, and advantages of the CUT&Tag method, and discusses its potential to advance the frontiers of life science research.
What Is CUT&Tag?
CUT&Tag is a highly sensitive epigenomic profiling technique that enables precise labeling and cleavage of chromatin regions by targeting chromatin-associated proteins. This allows for efficient mapping of histone modifications and transcription factor binding sites. Compared with ChIP-seq, CUT&Tag offers superior sensitivity, reduced background noise, and minimal input requirements, making it particularly advantageous for studies involving rare cell populations or limited starting material. Currently, it has been widely applied in oncology, immunology, developmental biology, and neuroscience research.
Principles and Mechanistic Basis of CUT&Tag
The fundamental mechanism of CUT&Tag involves antibody-mediated precise cleavage and tagging of chromatin regions bound to specific chromatin-associated proteins. The process can be summarized in three key steps:
1. Specific Antibody Recognition of the Target Protein
Following mild fixation and permeabilization of cells or tissue samples, a specific antibody is introduced to bind the target chromatin-associated protein (e.g., histone modifications, transcription factors). This step ensures the accuracy and specificity of subsequent reactions.
2. Recruitment of the Protein A/G–Tn5 Transposase Fusion Complex
A fusion protein consisting of Protein A or Protein G conjugated to Tn5 transposase is added. These fusion proteins specifically recognize and bind to the Fc region of the antibody, thereby localizing the transposase to chromatin regions in close proximity to the target protein binding sites.
3. Tn5 Transposase-Mediated DNA Cleavage and Adapter Integration
In the presence of Mg²⁺, the Tn5 transposase becomes catalytically active, cleaving chromatin DNA fragments while simultaneously inserting sequencing adapter sequences at the cleavage sites. These adapter-tagged DNA fragments can then be amplified via PCR to construct sequencing libraries for high-throughput analysis.
Experimental Workflow of CUT&Tag
Compared with ChIP-seq, the CUT&Tag procedure is more streamlined and time-efficient. The principal steps are as follows:
1. Sample Preparation
(1) Collect cells, with or without mild fixation.
(2) Permeabilize cells to facilitate antibody access to nuclear chromatin.
2. Incubation with Specific Antibody
The sample is incubated with the specific antibody at 4 °C for several hours to overnight to ensure sufficient binding.
3. Incubation with Protein A/G–Tn5 Complex
The Protein A/G–Tn5 fusion complex is introduced and incubated for approximately 1 hour, enabling precise localization to the target protein vicinity.
4. Chromatin Fragmentation and Adapter Tagging
The addition of Mg²⁺ activates the Tn5 transposase, allowing DNA cleavage and adapter integration within several minutes.
5. DNA Recovery and Library Preparation
(1) DNA fragments are directly released, eliminating the need for complex immunoprecipitation or purification procedures.
(2) PCR amplification is performed, followed by high-throughput sequencing.
Advantages and Applications of CUT&Tag
1. Key Advantages
(1) High sensitivity: CUT&Tag can be performed with as few as 100–1000 starting cells and is applicable even at the single-cell level.
(2) Low background noise: The absence of immunoprecipitation steps minimizes nonspecific signals, yielding data with improved signal-to-noise ratios.
(3) Operational efficiency: The entire procedure can be completed within 1–2 days, substantially increasing experimental throughput.
2. Broad Applicability
The high sensitivity, specificity, and throughput of CUT&Tag make it suitable for a wide range of research contexts, including:
(1) Epigenetic profiling of rare cell populations (e.g., stem cells or cancer stem cells).
(2) High-resolution mapping of chromatin modifications to investigate disease-associated epigenetic alterations.
(3) Single-cell chromatin analysis to study cellular heterogeneity and dynamic regulatory processes.
CUT&Tag vs. ChIP-seq: Considerations for Method Selection
While CUT&Tag offers multiple advantages, the choice between CUT&Tag and ChIP-seq should be guided by the experimental requirements.
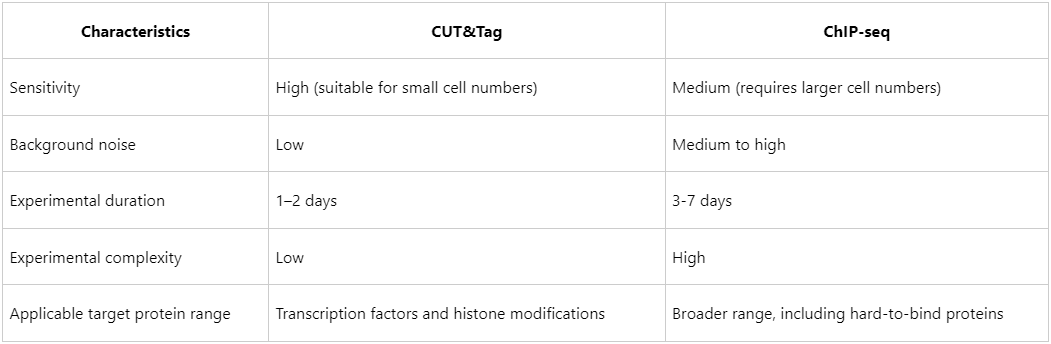
For studies with low-input samples or requiring rapid, high-sensitivity results, CUT&Tag is often preferable. Conversely, for more complex protein targets, ChIP-seq may remain the more robust option.
The growing adoption of CUT&Tag in proteomics and chromatin epigenetics underscores its transformative potential. Nonetheless, successful implementation depends on high-quality antibodies, Tn5 transposase, and optimized protocols. Commercial suppliers such as MtoZ Biolabs offer high-purity antibodies, precisely engineered Protein A/G–Tn5 fusion enzymes, and comprehensive technical support, facilitating accurate and efficient chromatin profiling. With its exceptional sensitivity and operational simplicity, CUT&Tag is poised to supplement or replace ChIP-seq in many applications, driving future breakthroughs in epigenetic research.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







