Tyrosine Phosphorylation Proteomics Analysis Service
- Tyrosine Phosphorylation Site Identification: Comprehensive mapping of tyrosine phosphorylation sites on proteins across complex biological samples, from cell lines to tissues.
- Quantification of Tyrosine Phosphorylation Levels: Accurate quantification of phosphorylation events to track dynamic changes in response to treatments or disease conditions.
- Pathway Analysis: Integration of tyrosine phosphorylation data with known signaling pathways to understand the functional implications of phosphorylation in cellular responses.
- Post-translational Modification Profiling: Investigation of tyrosine phosphorylation in combination with other PTMs, such as ubiquitination or acetylation, to provide a more holistic view of cellular regulation.
- Signal Transduction Pathway Analysis: Mapping tyrosine phosphorylation networks involved in cell signaling, including growth factor, cytokine, and immune response pathways.
- Drug Development: Investigating the role of tyrosine kinases in diseases, aiding in the design of tyrosine kinase inhibitors and other targeted therapies for cancer and other diseases.
- Cellular Mechanisms: Investigating the role of tyrosine phosphorylation in essential cellular processes such as cell division, migration, and differentiation.
- Disease Mechanisms: Understanding how dysregulated tyrosine phosphorylation contributes to various diseases, including metabolic disorders, cardiovascular diseases, and immune dysfunction.
Tyrosine Phosphorylation Proteomics is an advanced analytical technique designed to identify and quantify tyrosine phosphorylation sites within the proteome. Tyrosine phosphorylation occurs when a phosphate group is covalently attached to the hydroxyl group of the tyrosine residue within a protein, catalyzed by enzymes called tyrosine kinases. Although the proportion of tyrosine phosphorylation is relatively small compared to serine/threonine phosphorylation, it plays a crucial role in regulating various cellular processes. Tyrosine phosphorylation is vital for controlling protein-protein interactions and initiating signaling cascades that mediate cellular responses to growth factors, cytokines, and other extracellular signals. This modification is closely associated with the regulation of cell fate and is critical in disease mechanisms, particularly in cancer, where tyrosine kinases account for 75% of oncogenes, and 63% of protein kinase-targeted cancer therapies are tyrosine kinase inhibitors. Therefore, tyrosine phosphorylation proteomics analysis provides invaluable insights into disease mechanisms and therapeutic potential.
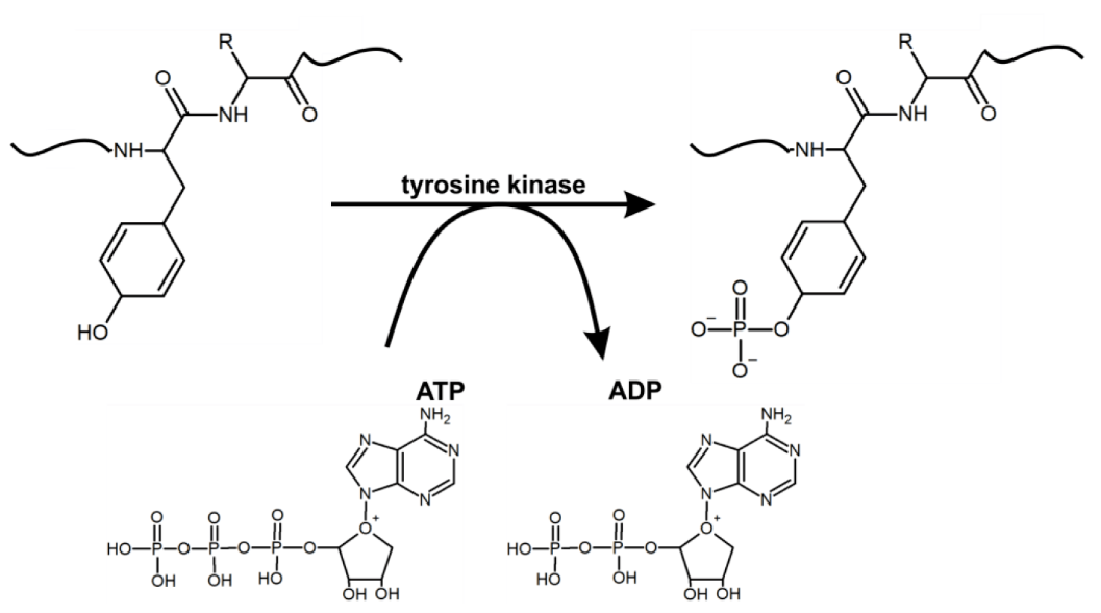
Figure 1. The Mechanism of Tyrosine Residue Phosphorylation in Proteins
Service at MtoZ Biolabs
MtoZ Biolabs offers a comprehensive Tyrosine Phosphorylation Proteomics Analysis Service to identify and quantify tyrosine phosphorylation sites with high resolution and sensitivity. Our Tyrosine Phosphorylation Proteomics Analysis Service employs cutting-edge liquid chromatography-tandem mass spectrometry (LC-MS/MS) technology combined with selective enrichment methods, like phosphopeptide enrichment using IMAC (immobilized metal affinity chromatography) or TiO₂ (titanium dioxide) methods, to maximize the sensitivity and specificity of tyrosine phosphorylation site identification.
MtoZ Biolabs' Services include:
Analysis Workflow
1. Sample Preparation: The first step involves protein extraction from the biological sample (e.g., cultured cells, tissue) and subsequent digestion into peptides using enzymes like trypsin.
2. Phosphopeptide Enrichment: Tyrosine-phosphorylated peptides are enriched using advanced techniques such as IMAC or TiO₂ chromatography to selectively capture phosphopeptides from the complex sample.
3. LC-MS/MS Analysis: The enriched peptides are analyzed using high-resolution LC-MS/MS, where peptides are separated by liquid chromatography and then analyzed by tandem mass spectrometry to identify the tyrosine phosphorylation sites.
4. Data Analysis & Bioinformatics Interpretation: Using specialized software and bioinformatics tools, the resulting data is analyzed to identify specific phosphorylation sites, quantify phosphorylation levels, and map the phosphoproteome across the sample. The identified phosphorylation sites are mapped to cellular signaling pathways to understand their biological relevance and potential therapeutic implications.

Figure 2. Workflow for Tyrosine Phosphorylation Proteomics Analysis Service
Service Advantages
✅High Sensitivity: MtoZ Biolabs ensures precise detection of low-abundance tyrosine phosphoproteins, even in complex biological samples.
✅Expert Team: Our team of experienced scientists specializes in post-translational modification analysis and provides valuable insights into the mechanisms behind your research.
✅Comprehensive Analysis: MtoZ Biolabs offers end-to-end solutions, from sample preparation to data analysis, ensuring you have a complete picture of tyrosine phosphorylation in your samples.
✅Customized Solutions: Our Tyrosine Phosphorylation Proteomics Analysis Services are tailored to meet the specific needs of your research, whether you are studying basic biology, disease mechanisms, or drug development.
Applications
MtoZ Biolabs' Tyrosine Phosphorylation Proteomics Analysis Service is widely used in various research and therapeutic applications:
Case Study
Tyrosine Phosphorylation Proteomics Analysis in Acute Myeloid Leukemia (AML)
Tyrosine phosphorylation plays a crucial role in cancer progression by regulating key signaling pathways. In a recent study, a deep-scale proteome and phosphoproteome analysis was conducted on 44 AML patient samples and six healthy controls to identify dysregulated tyrosine phosphorylation events in leukemia pathogenesis. Through mass spectrometry-based tyrosine phosphorylation proteomics analysis, researchers detected nearly 30,000 phosphosites, including significantly increased phosphorylation of tyrosine residues in key regulatory proteins such as PTPN11 (Tyr-542), PRKCD (Tyr-313), and STAT3 (Tyr-705). Notably, these phosphorylation events were observed in AML samples but were absent or significantly reduced in healthy bone marrow controls, indicating their potential as biomarkers and therapeutic targets. Further analysis revealed that phosphorylation of STAT3 Tyr-705, a site critical for transcriptional activity, was highly enriched in AML, reinforcing its role in leukemia progression. This study underscores the power of Tyrosine Phosphorylation Proteomics Analysis Service in identifying disease-associated phosphorylation patterns. Tyrosine Phosphorylation Proteomics Analysis Service aids researchers in elucidating signaling dysregulation in cancer, facilitating biomarker discovery, and supporting drug development for targeted therapies.
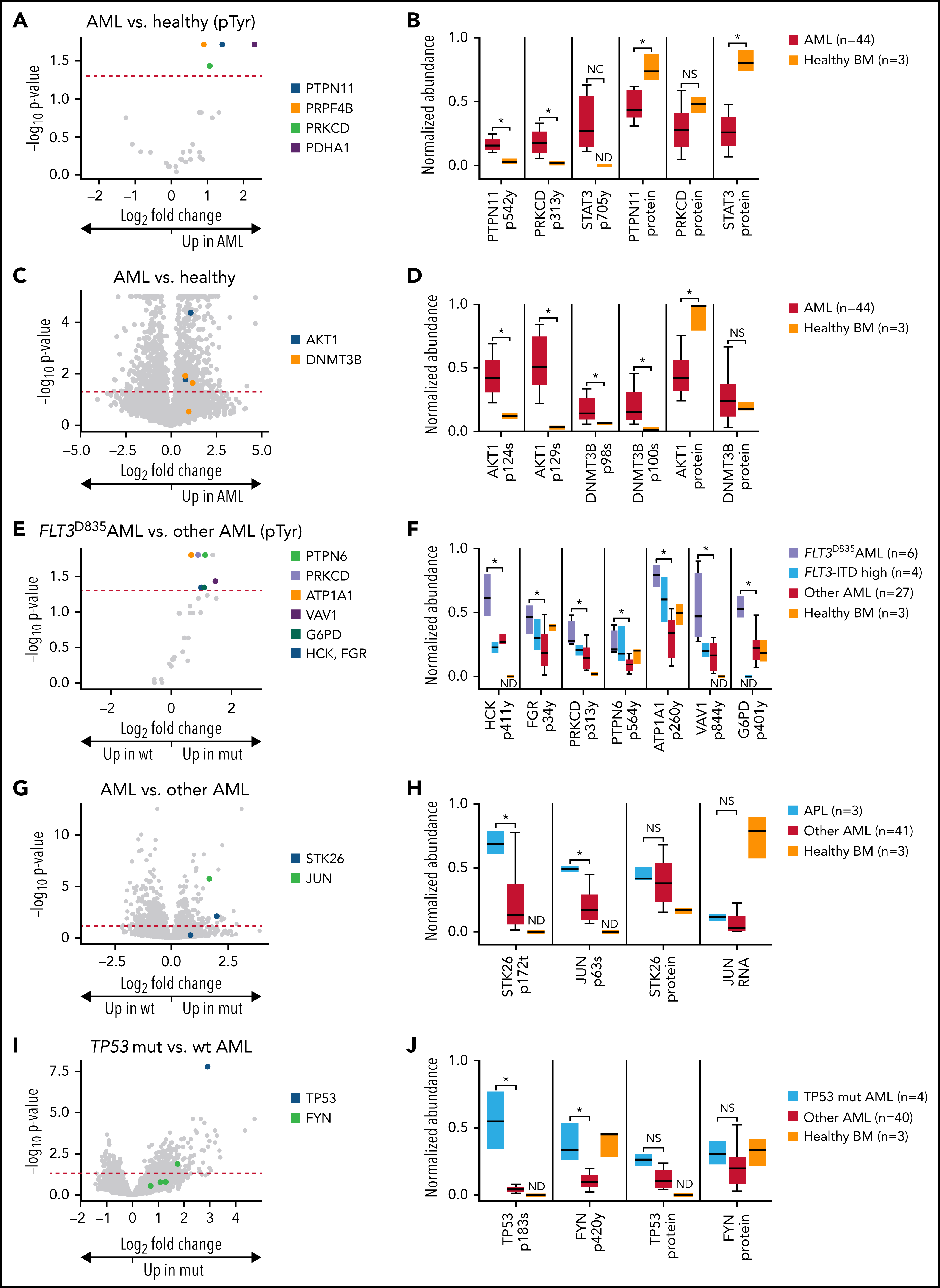
Figure 3. Phosphoproteomic Analyses of AML Samples Associated with Apecific Mutations
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
Contact us today to explore how our Tyrosine Phosphorylation Proteomics Analysis Service can accelerate your research and therapeutic development!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Tyrosine Phosphoproteomics Analysis Service
How to order?







