Tumor Phosphorylation Signaling Pathway Analysis Service
- Personalized Therapy Design: Identifies hyperactive kinases or dysregulated pathways in tumors to guide targeted therapy selection and combination strategies.
- Drug Target Discovery: Pinpoints actionable phosphorylation-driven oncogenic targets for small-molecule inhibitor or monoclonal antibody development.
- Biomarker Development: Discovers phosphorylation-specific biomarkers for early cancer detection, treatment response monitoring, and recurrence prediction.
- Resistance Mechanism Elucidation: Maps adaptive phosphorylation signaling rewiring in drug-resistant tumors to design resistance-overcoming therapeutic regimens.
- Prognostic Stratification: Correlates phosphoprotein activation patterns with clinical outcomes to refine risk assessment and survival prediction models.
- Pathway Crosstalk Analysis: Deciphers interactions between phosphorylation cascades (e.g., MAPK, PI3K/AKT) and other oncogenic signaling networks (e.g., hypoxia, immune checkpoints).
Tumor phosphorylation signaling pathway analysis is an advanced analytical approach that focuses on mapping and understanding the phosphorylation events within signaling pathways in cancer cells. Phosphorylation, a critical post-translational modification, regulates various cellular processes, including cell growth, survival, and metabolism. Phosphorylation governs the activation and deactivation of key signaling pathways, such as those involving EGFR, MAPK, PI3K/AKT, and JAK/STAT. These pathways control tumor progression and therapy response, with dysregulated phosphorylation contributing to oncogenesis and therapy resistance. Analyzing tumor-specific phosphorylation signatures helps differentiate cancer cells from normal cells, providing insights into the molecular mechanisms of disease and therapeutic response.
Tumor phosphorylation signaling pathway analysis is essential for unraveling the molecular events driving cancer. Tumor Phosphorylation Signaling Pathway Analysis enables identification of crucial phosphorylation sites, elucidates how signaling networks are altered in response to external factors, and uncovers novel biomarkers and therapeutic targets. By profiling these signaling pathways in tumors, researchers can gain deeper insight into cancer biology and potential resistance mechanisms.
Service at MtoZ Biolabs
MtoZ Biolabs provides a Tumor Phosphorylation Signaling Pathway Analysis Service that offers a comprehensive approach to understanding phosphorylation-mediated signaling networks in cancer. Utilizing advanced mass spectrometry-based phosphoproteomics, we enable precise identification and quantification of phosphorylation events that regulate tumor progression, therapeutic response, and resistance mechanisms. Our Tumor Phosphorylation Signaling Pathway Analysis Service platform integrates high-resolution LC-MS/MS phosphoproteomics, multiple phosphopeptide enrichment strategies and quantitative proteomics techniques such as label-free quantification (LFQ), stable isotope labeling (SILAC), and TMT/iTRAQ labeling. This enables deep coverage of phosphoproteomes, providing site-specific phosphorylation mapping, dynamic pathway profiling, and comparative analysis across different tumor samples.
Analysis Workflow
1. Sample Preparation: Extract proteins from tumor tissues or cell lines, ensuring optimal preservation of phosphorylation states.
2. Protein Digestion: Enzymatic digestion of proteins into peptides for phosphoproteomic analysis.
3. Phosphopeptide Enrichment: Enrich phosphorylated peptides using affinity-based, MOAC, IMAC, or alternative strategies.
4. LC-MS/MS Analysis: High-resolution mass spectrometry for phosphopeptide identification and quantification.
5. Data Processing: Perform raw data processing, peak alignment, and normalization to ensure accuracy.
6. Pathway and Functional Analysis: Map phosphorylation sites to signaling pathways, kinases, and functional networks.
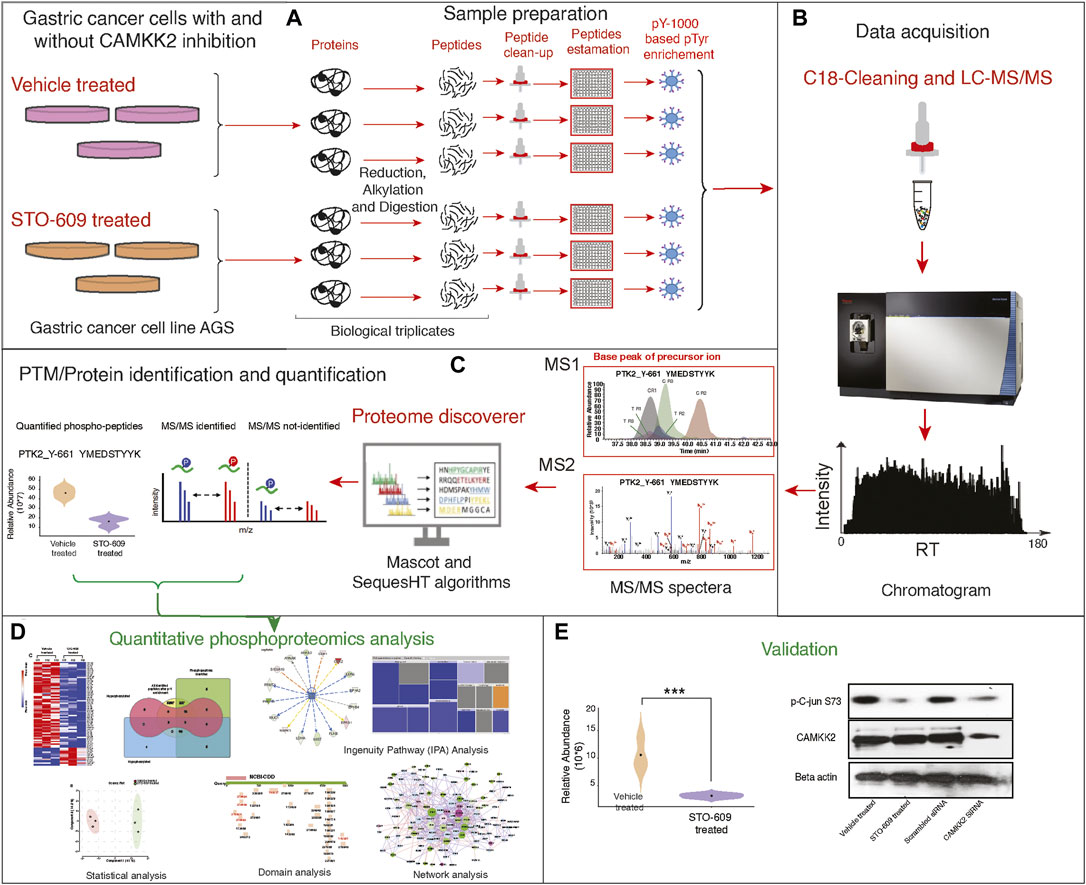
Figure 1. Detailed Experimental Workflow for Phospho-(Y)-Proteomic Analysis, Data Analysis, and Functional Validation in Gastric Cancer Cells
Service Advantages
✅Expertise in Phosphoproteomics: Our team consists of highly skilled professionals with deep knowledge of tumor phosphorylation signaling pathways, ensuring the highest level of technical proficiency and scientific insight in every analysis.
✅Advanced Platform: We utilize state-of-the-art mass spectrometry platforms and advanced enrichment techniques, providing precise and comprehensive phosphorylation analysis across complex biological samples.
✅Customized Solutions: Our Tumor Phosphorylation Signaling Pathway Analysis Service is fully tailored to meet the unique needs of each project, offering personalized workflows to address specific research goals, from basic mechanistic studies to drug target discovery.
✅Rapid Turnaround: MtoZ Biolabs offers fast, efficient service with quick sample processing and data analysis.
Applications
Our Tumor Phosphorylation Signaling Pathway Analysis Service is applicable in various sectors:
Case Study
Tumor Phosphorylation Signaling Pathway Analysis in Metastatic Colorectal Cancer (mCRC)
In a recent study, mass spectrometry-based proteomics and phosphoproteomics were employed to explore the molecular basis of response to EGFR blockade in metastatic colorectal cancer (mCRC) patient-derived xenografts (PDXs). This study aimed to identify distinct phosphorylation signatures associated with drug sensitivity and resistance. The results showed significant differences in the phosphorylation profiles between sensitive and resistant tumors. Sensitive tumors exhibited increased pathway activity related to MAPK inhibition and abundant tyrosine phosphorylation of cell junction proteins like CXADR and CLDN1/3. In contrast, resistant tumors were characterized by epithelial-mesenchymal transition, as well as elevated MAPK and AKT signaling. Additionally, the study identified hyperactive kinases in cetuximab-resistant PDX models, including members of the Src and ephrin kinase families, which were linked to unexplained resistance. Targeting these hyperactive kinases, either alone or in combination with cetuximab, led to growth inhibition of ex vivo organoids and in vivo PDX models. This analysis underscores the value of Tumor Phosphorylation Signaling Pathway Analysis Service in understanding treatment responses and identifying alternative drug targets in resistant tumors.
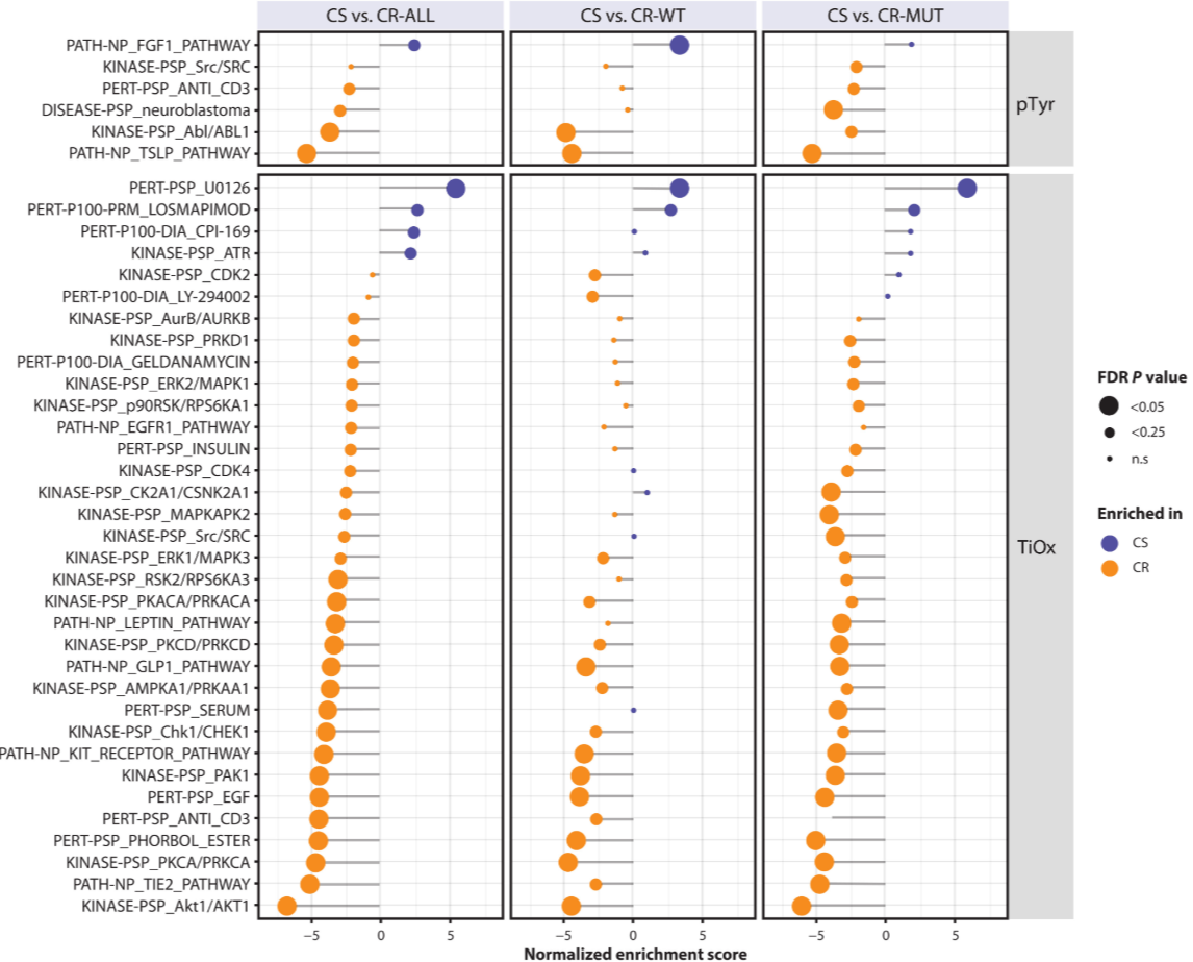
Figure 2. Posttranslational Modification Signature Enrichment Analysis of Cetuximab-Sensitive and Cetuximab-Resistant Biology
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
By elucidating oncogenic signaling networks, MtoZ Biolabs helps researchers gain critical insights into phosphorylation-driven tumor processes. Our Tumor Phosphorylation Signaling Pathway Analysis Service is designed to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Phosphoproteomics Service
How to order?







