Top 5 Methods for Protein Structure Analysis and Their Applications
The function of a protein is intrinsically linked to its three-dimensional structure. Protein structure analysis not only facilitates the interpretation of biological behavior but is also extensively utilized in diverse research areas, including drug discovery, antibody engineering, and the elucidation of metabolic pathways. Currently, researchers have access to multiple analytical techniques for analyzing protein structure, each with distinct advantages, limitations, and suitability for specific protein types and research objectives.
X-ray Crystallography
1. Methods
X-ray crystallography is a high-resolution structural determination technique. By analyzing the diffraction patterns of protein crystals exposed to X-rays, electron density maps are generated, enabling reconstruction of the atomic arrangement. This method can achieve sub-0.8 Å resolution, providing atomic-level structural insights.
2. Applications
(1) High-resolution structural modeling: Ideal for generating accurate three-dimensional models of monomeric proteins and protein complexes.
(2) Structure-based drug design: Facilitates small-molecule screening and optimization guided by structural data.
(3) Functional mechanism elucidation: Enables detailed analysis of catalytic sites, active center conformations, and other critical structural features.
3. Considerations
The method requires exceptionally high-quality crystals, and certain classes of proteins, such as membrane proteins and large complexes, remain challenging to crystallize.
Cryo-Electron Microscopy (Cryo-EM)
1. Methods
Cryo-EM employs an electron beam as the imaging source to capture and reconstruct three-dimensional structures of rapidly vitrified protein particles. The method eliminates the need for crystallization and requires minimal sample quantities, establishing itself as an increasingly important approach in structural biology.
2. Applications
(1) Structural characterization of large macromolecular assemblies: Particularly effective for ribosomes, viral particles, and other high-molecular-weight systems.
(2) Membrane protein studies: Overcomes limitations associated with crystallographic methods for membrane-associated proteins.
(3) Analysis of conformational heterogeneity: Allows visualization of proteins across multiple functional states.
3. Considerations
The technique necessitates highly homogeneous samples and robust capabilities in data acquisition and image processing, making it more suitable for research teams with established expertise.
Nuclear Magnetic Resonance (NMR)
1. Methods
NMR spectroscopy utilizes the resonance behavior of nuclear spins in a magnetic field to determine interatomic distances and connectivity, thereby reconstructing the three-dimensional structure of proteins in solution. It is particularly advantageous for probing dynamic aspects of protein behavior.
2. Applications
(1) Small protein structure analysis: Most suitable for proteins with molecular weights below 30 kDa.
(2) Investigation of dynamic behavior: Captures conformational exchanges, flexible domains, and intrinsically disordered regions.
(3) Protein-ligand interaction studies: Identifies binding sites and conformational adaptations upon ligand engagement.
3. Considerations
As molecular weight increases, spectral complexity and data acquisition become increasingly challenging, necessitating advanced instrumentation and meticulous sample preparation.
Cross-Linking Mass Spectrometry (XL-MS)
1. Methods
XL-MS employs chemical cross-linkers to covalently link proximal amino acid residues within or between proteins. The cross-linked peptides are then detected by mass spectrometry to infer spatial proximities, imposing distance restraints that inform the overall structural model.
2. Applications
(1) Coarse-grained conformational modeling: Provides spatial constraints to guide three-dimensional structural modeling.
(2) Protein interaction mapping: Identifies key inter- and intramolecular contact regions.
(3) Corroboration of other structural data: Frequently used to complement Cryo-EM and X-ray crystallography models.
3. Considerations
The technique offers relatively low resolution and is generally applied as a supplementary or high-throughput screening tool rather than as a standalone method for constructing detailed structural models.
Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS)
1. Methods
HDX-MS monitors the exchange of backbone amide hydrogens with deuterium in heavy water, thereby assessing solvent accessibility and conformational stability across different protein regions. This provides dynamic structural information.
2. Applications
(1) Tracking conformational dynamics: Suitable for studying protein activation, folding, and stability transitions.
(2) Antigen-antibody epitope mapping: Widely employed in antibody engineering and biopharmaceutical development.
(3) Protein engineering optimization: Identifies flexible regions to support the design of more stable protein variants.
3. Considerations
The method is highly sensitive, with data interpretation reliant on high-resolution mass spectrometry and sophisticated computational analysis.
Recommendations for Method Selection
Each protein structure analysis technique differs in resolution, dynamic capabilities, sample requirements, and throughput. Optimal outcomes are often achieved by strategically combining methods according to specific scientific objectives.
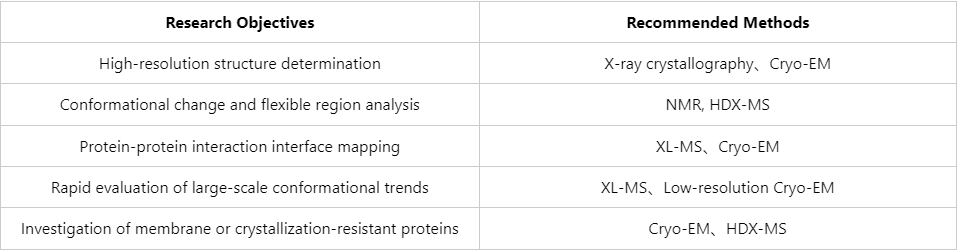
With the increasing demand for detailed protein structural information in biopharmaceuticals and synthetic biology, researchers face challenges such as sample complexity, diverse experimental targets, and stringent timelines. Selecting appropriate structural determination strategies, combined with rigorous sample handling and comprehensive data analysis, is essential for successful outcomes. MtoZ Biolabs leverages expertise in proteomics and mass spectrometry, alongside structural biology applications, to deliver protein structure analysis services. Our mission is to advance fundamental research and technological translation through collaborative studies of protein structure and function.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







