The Role of Phosphoproteomics in Multi-Omics Integration
In the post-genomic era, life science research is transitioning from isolated, single-layer investigations toward comprehensive systems-level integration. Technologies such as transcriptomics, proteomics, metabolomics, epigenomics, and single-cell omics each provide snapshots of biological systems at distinct molecular layers. However, only through multi-omics integration can the dynamic and coordinated nature of biological processes be fully reconstructed. Within this integrative framework, phosphoproteomics plays a pivotal intermediary role. It serves as a functional link between gene expression and biological activity, while also capturing rapid cellular responses to external stimuli and underlying signaling regulatory mechanisms. As one of the most prevalent and functionally important post-translational modifications, protein phosphorylation directly regulates key biological processes, including signal transduction, metabolic pathways, and cell-cycle control. In multi-omics studies, the integrated analysis of phosphoproteomics with genomic, transcriptomic, and metabolomic data enables not only the elucidation of how genetic alterations propagate through signaling networks to modulate protein activity, but also the identification of critical regulatory sites and their functional consequences. Such integrative strategies are essential for dissecting complex biological systems, uncovering disease mechanisms, and facilitating drug target discovery.
Phosphoproteomics in the Omics Puzzle: What Is Its Unique Value?
1. Temporal Dimension: The Earliest Indicator of Cellular State Changes
Unlike genetic mutations or transcriptional up- or downregulation, phosphorylation is a rapid and reversible form of post-translational regulation that typically occurs within seconds to minutes following pathway activation. It provides an immediate readout of cellular responses to:
(1) Drug stimulation
(2) Stress responses
(3) Growth factor signaling
(4) Inflammatory or immune activation
Consequently, in multi-omics investigations, phosphoproteomics represents one of the earliest molecular layers to exhibit dynamic changes, offering high temporal resolution for studying regulatory events.
2. Mechanistic Dimension: Identifying Functional Switches within Signaling Pathways
In canonical signaling pathways such as PI3K/AKT/mTOR, MAPK, and JAK–STAT, the activity of most key components, including kinases and transcription factors, is governed by changes in phosphorylation status. While transcriptomic analyses capture variations in mRNA abundance, they are unable to directly reflect protein functional states. Phosphoproteomics enables the identification of regulatory nodes within signaling cascades that are modulated by pharmacological interventions and therefore constitutes a central tool for mechanistic studies.
3. System Dimension: Intrinsic Compatibility with Other Omics Layers
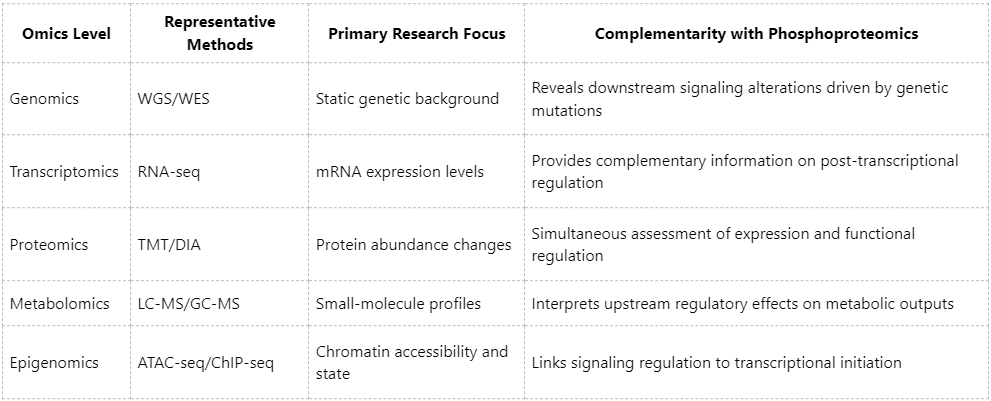
Representative Applications in Multi-Omics Integrative Studies
1. Dissection of Tumor Signaling Pathways
In cancer research, transcriptomic data alone are often insufficient to predict drug sensitivity. For instance, although multiple tumor subtypes may exhibit comparable expression levels of PI3K pathway genes, only those with significantly elevated p-AKT levels respond effectively to AKT inhibitors.
Integrative strategy:
(1) Transcriptomics: assess baseline expression of pathway components.
(2) Phosphoproteomics: determine kinase activity and pathway activation status.
(3) Targeted metabolomics: evaluate downstream metabolic alterations.
(4) Outcome: establish a comprehensive model linking signal regulation → phenotypic output.
2. Drug Screening and Mechanism-of-Action Analysis
During the development of small-molecule or antibody-based therapeutics, it is critical to determine which signaling pathways are modulated and which biological processes are affected.
Integrative strategy:
(1) Following drug treatment, perform combined phosphoproteomic, proteomic, and metabolomic analyses.
(2) Systematically evaluate drug efficacy by tracing changes from kinase activity to downstream effector regulation and cellular metabolic states.
3. Clinical Molecular Subtyping and Biomarker Discovery
Phosphoproteomics is frequently employed to support molecular classification, for example:
(1) Stratifying tumors of the same histological type into subgroups based on phosphorylation patterns such as p-ERK or p-AKT.
(2) Integrating phosphorylation data with expression and mutation profiles to construct highly predictive multi-omics biomarker models.
Data Integration Strategies: Enabling Phosphoproteomics to Convey Biological Meaning
1. Pathway-Oriented Hierarchical Integration
By leveraging curated databases such as Reactome and KEGG, signaling pathways can be defined and multi-omics data mapped onto structured pathway frameworks:
(1) Identification of activated pathways based on significant phosphorylation changes.
(2) Detection of pathway components with aberrant mRNA or protein expression.
(3) Assessment of accumulated downstream metabolites.
This approach facilitates an integrated, systems-level understanding of biological regulation.
2. Network Analysis Combined with Machine Learning
Multi-omics regulatory networks can be constructed by integrating:
(1) Kinase–substrate relationship prediction (PhosphoSitePlus combined with motif analysis).
(2) Protein–protein interaction networks (STRING).
(3) Multidimensional feature extraction and classification modeling (e.g., LASSO, SVM, RF).
Such frameworks support disease subtype classification, prediction of drug responses, and identification of key regulatory nodes.
Phosphoproteomics is not merely an extension of functional proteomics but a central signal-decoding component within multi-omics research. By capturing dynamic functional states, it transforms static gene expression measurements into biologically actionable insights. In future studies of disease mechanisms, drug development, and precision medicine, only by fully integrating the continuum from gene → expression → regulation → phenotype can the fundamental complexity of biological systems be comprehensively understood. For researchers planning multi-omics studies with a focus on signaling pathways, kinase activity, or dynamic regulatory mechanisms, MtoZ Biolabs offers system-level analytical perspectives and engineered platforms to support advanced research initiatives.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







