Strengths and Limitations of DIA in Protein Quantification
-
Overlapping and interfering fragment peaks, which complicate spectral deconvolution;
-
Increased dependence on the accuracy of identification algorithms and the quality of spectral libraries, as the matching process is less straightforward than the one-to-one precursor-fragment pairing in DDA.
-
Implementing optimized windowing schemes (e.g., dynamic or variable-width windows) to minimize co-fragmentation;
-
Generating high-quality, high-coverage project-specific spectral libraries;
-
Employing advanced algorithms such as DIA-NN, Spectronaut, and FragPipe to enhance identification accuracy.
-
Raw data preprocessing (peak detection and calibration)
-
Peptide-level matching and quantification (spectral deconvolution)
-
Protein-level summarization and normalization
-
Statistical analysis, differential testing, and biological interpretation
-
Statistical tests for differential proteins (e.g., t-tests, ANOVA)
-
GO/KEGG functional annotation and enrichment analysis
-
Protein interaction network construction and clustering analysis
-
Publication-ready figures tailored for SCI journal submissions
-
Post-translational modification (PTM) studies (e.g., phosphorylation, acetylation): the low abundance of modified peptides and severe co-fragmentation significantly increase analytical complexity;
-
Quantification of ultra-low abundance proteins: even with comprehensive acquisition, the signals of some proteins remain difficult to detect reliably;
-
Identification of novel or mutant peptides: this demands exceptionally comprehensive spectral libraries and robust search algorithms, often necessitating complementary DDA-based analysis.
As proteomics continues to advance in both depth and breadth, studies involving numerous samples, varying conditions, and complex phenotypes increasingly demand stability, coverage, and reproducibility in protein quantification. Although Data-Dependent Acquisition (DDA) has been widely employed in proteomic research, its limitations—such as detection bias, poor reproducibility, and the frequent omission of low-abundance proteins—have become more apparent with growing study complexity. The emergence of Data-Independent Acquisition (DIA) offers a promising alternative, enabling robust, high-throughput, and reproducible protein quantification. This paper systematically examines the strengths, bottlenecks, and applicability of DIA in protein quantification.
Core Mechanism of DIA for Protein Quantification
DIA mass spectrometry operates by segmenting the full precursor ion m/z range into a series of consecutive windows. Within each window, all precursor ions are simultaneously fragmented, enabling comprehensive and unbiased ion sampling.
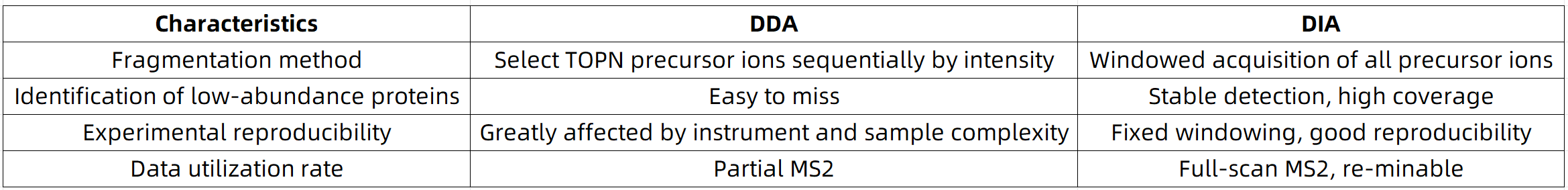
DIA vs. DDA Acquisition
Key Advantages of DIA in Protein Quantification
1. Increased Protein Detection Coverage
Data-dependent acquisition (DDA) fragments only the top N most intense precursor ions, making it prone to missing signals, especially in complex samples or those rich in low-abundance proteins. In contrast, data-independent acquisition (DIA) systematically and comprehensively collects fragment ions across all precursors, significantly enhancing:
(1) The overall depth of proteome coverage
(2) The sensitivity toward low-abundance and regulatory proteins
(3) The consistency of detection across biological replicates
For instance, in complex biological matrices such as tissue, plasma, or FFPE samples, DIA frequently identifies 6000 to 8000 proteins—markedly outperforming DDA under comparable conditions.
2. Improved Reproducibility and Stability in Quantification
DIA employs a fixed acquisition scheme with predetermined windows, mitigating the stochastic peptide sampling seen in DDA due to ion intensity variability. This consistent acquisition yields several advantages:
(1) Lower coefficients of variation among biological replicates (CV < 15%)
(2) Enhanced suitability for integrative multi-omics and clinical cohort studies
(3) Compatibility with high-throughput automation and AI-driven analysis
In large-scale DIA studies conducted by MtoZ Biolabs, over 90% of proteins were consistently identified and quantified across more than 20 samples, fulfilling statistical rigor for biomedical research.
3. Supports Both Library-Based and Library-Free Quantification
DIA data analysis can follow two complementary approaches:
(1) Library-based analysis: Employs a pre-established spectral library for matching, achieving higher quantitative precision;
(2) Library-free (DirectDIA) analysis: Suitable for species or sample types lacking prior data, relying on advanced algorithms to directly interpret DIA spectra.
This flexibility ensures that DIA remains broadly applicable—even in studies involving novel samples, non-model organisms, or constrained replicates.
4. Reusability and Long-Term Value of Data
DIA records comprehensive MS/MS fragment data, enabling future reinterpretation and analysis, such as:
(1) Re-searching unidentified peptides using updated spectral libraries
(2) Expanding quantitative targets to newly discovered proteins
(3) Integrating proteomic data with later transcriptomic or metabolomic datasets
In contrast, DDA's real-time precursor selection limits retrospective analysis and recovery of missed signals. As such, DIA offers superior long-term data utility, particularly valuable for longitudinal studies and platform-scale research initiatives.
Limitations of DIA in Protein Quantification
Despite its numerous advantages, DIA still presents certain technical bottlenecks and challenges that should be carefully considered during experimental design and data interpretation.
1. High Spectral Complexity and Fragment Ion Interference Complicating Peptide Identification
DIA fragments all precursor ions within a given window simultaneously, resulting in MS2 spectra that contain a mixture of fragment ions from multiple peptides. This introduces:
Mitigation strategies include:
2. Complex Data Processing Workflow and High Analytical Barrier
DIA data analysis involves multiple sequential steps, including:
Compared to DDA, this workflow is more intricate and imposes greater demands on computational proficiency and data interpretation expertise. To address these challenges, MtoZ Biolabs offers an integrated bioinformatics solution, encompassing:
3. Limitations in Specific Research Contexts
Although DIA is applicable to the majority of proteomic quantification studies, certain scenarios still present analytical limitations:
Protein Quantification Using DIA: Services Provided by MtoZ Biolabs
MtoZ Biolabs, a dedicated provider of high-resolution mass spectrometry and proteomics services, has developed a robust and comprehensive data-independent acquisition (DIA) platform for protein quantification, featuring:
1. Multi-instrument compatibility: supporting Thermo Orbitrap Exploris, Bruker timsTOF, SCIEX TripleTOF, among others.
2. End-to-end standardized workflow: encompassing sample preparation, enzymatic digestion, DIA data acquisition, spectral library generation, and downstream data analysis.
3. Customizable project strategies: enabling flexible selection of approaches such as label-free DIA or TMT-DIA according to specific research goals.
4. Standardized bioinformatics output: all results are delivered with publication-ready figures and analysis reports suitable for submission to SCI-indexed journals.
We have collaborated successfully with a wide range of universities, hospitals, research institutes, and biopharmaceutical companies, gaining extensive practical experience in executing DIA-based protein quantification projects. These projects span diverse research areas, including oncology, immunology, neuroscience, and metabolism. As algorithmic performance improves and instrument capabilities advance, DIA is transitioning from a supplementary technique to a widely adopted mainstream approach. It is particularly advantageous for projects requiring comprehensive biological network profiling, high-throughput sample processing, and long-term data consistency. Nonetheless, a thorough understanding of DIA’s technical limitations, careful experimental design, and integration of expert data analysis remain critical to ensuring the reliability of results and successful publication. If you are planning a DIA-based study, MtoZ Biolabs offers expert consultation and technical support to help facilitate your research breakthroughs and enable the translation of findings into impactful outcomes.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







