SRM-MRM Quantitative Proteomics Services
SRM-MRM Quantitative Proteomics Services is a targeted protein quantification service based on triple quadrupole mass spectrometry, utilizing selected reaction monitoring (SRM) or multiple reaction monitoring (MRM) to achieve high specificity and high sensitivity quantification of proteins in samples. Its principle is to screen the characteristic peptides of the target protein through triple quadrupoles in series to ensure high selectivity and high sensitivity: the first quadrupole (Q1) selects specific precursor ions, the second quadrupole (Q2) fragments the parent ions into fragment ions through collision-induced dissociation (CID), and the third quadrupole (Q3) further screens specific fragment ions for detection and quantification. This precise ion selection mechanism enables SRM-MRM Quantitative Proteomics to achieve high-precision quantification of low-abundance proteins in complex biological samples such as plasma, tissue, and cerebrospinal fluid (CSF). Based on triple quadrupole mass spectrometry and liquid chromatography, MtoZ Biolabs provides SRM-MRM Quantitative Proteomics Services covering target protein screening, precise quantification, and bioinformatics analysis, supporting scientific research and clinical applications.
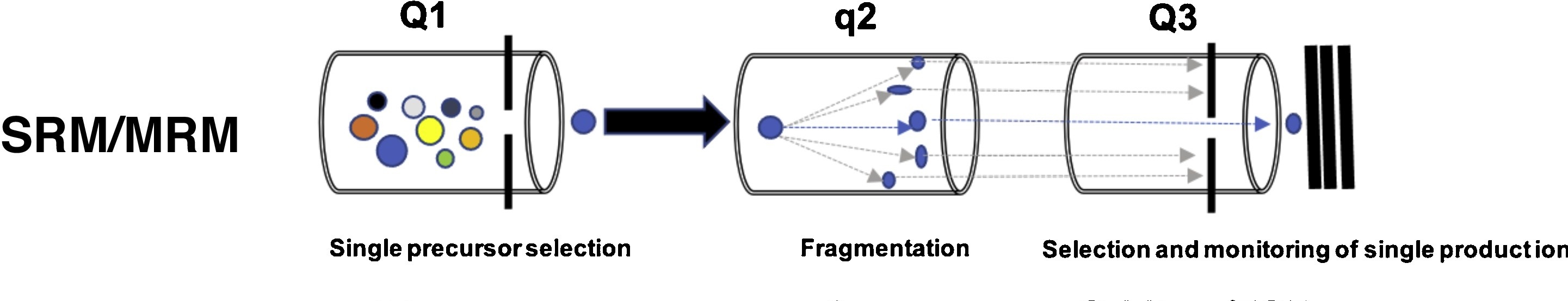
Li J. et al. Drug Discovery Today Technologies. 2021.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced SRM-MRM Quantitative Proteomics Services platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all SRM-MRM Quantitative Proteomics Services data, providing clients with a comprehensive data report.
Applications
Biomarker Validation
SRM-MRM Quantitative Proteomics can be used to validate potential biomarkers identified in non-targeted proteomics studies.
Protein Quantification Analysis
Enables absolute or relative quantification of specific proteins or peptides, suitable for detecting low-abundance proteins in complex biological samples.
Signaling Pathway Research
Quantifies key regulatory proteins to analyze the dynamic changes in signaling pathways.
Mechanism of Drug Action Studies
SRM-MRM Quantitative Proteomics can be used to assess the effects of drugs on target proteins or related signaling pathways.
Post-Translational Modification (PTM) Analysis
Combined with stable isotope labeling and other methods, it enables the quantitative analysis of PTM changes.
FAQ
Q. How to Optimize SRM-MRM Mass Spectrometry Parameters for Optimal Signal?
Optimizing SRM-MRM mass spectrometry parameters requires adjustments in multiple aspects, including ion selection, fragmentation optimization, and detection window:
1. Precursor Ion Selection: Choose high-response, stable m/z values for the target peptide while avoiding interference peaks.
2. Collision Energy (CE) Optimization: Adjust CE values for different peptides to achieve the most abundant and stable product ions.
3. Transition Optimization: Select high-abundance product ions with low background interference for accurate quantification.
4. Retention Time (RT) Stability: Optimize liquid chromatography gradient settings to ensure RT reproducibility and minimize drift.
5. Signal-to-Noise Ratio (S/N) Optimization: Set appropriate dwell time and integration time to improve detection sensitivity while minimizing noise interference.
Q. How to Analyze SRM-MRM Mass Spectrometry Data?
SRM-MRM data analysis mainly includes peak identification, quantitative calculation and data quality assessment. The specific process is as follows:
1. Data Import: Import mass spectrometry data using software such as Skyline, MultiQuant, and Pinpoint.
2. Peak Detection and Integration: Identify the characteristic peaks of the target peptide (Retention Time, RT) and ensure that the signal matches the preset ion pair (Transition).
3. S/N Evaluation: Apply an appropriate threshold (e.g., S/N > 10) to filter out low-confidence data for reliable quantification.
4. Quantitative Normalization:
External Standard Method: Calculate absolute quantification values based on a standard curve.
Internal Standard Method: Use stable isotope-labeled peptides to correct batch variations.
5. Data Quality Control (QC): Assess peak shape, retention time drift, and ion transition similarity (Dot Product, DotP) to ensure data consistency.
6. Statistical Analysis: Perform statistical tests to evaluate protein expression changes.
Finally, the processed data can be applied to protein quantification, post-translational modification (PTM) studies, and disease biomarker screening.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized SRM/MRM mass spectrometry to perform high-throughput targeted quantitative proteomic analysis of Labeo rohita, aiding in the establishment of a proteomic reference database for this species. The research presented quantitative expression profiles of multiple key proteins related to metabolism, immunity, and growth, providing high-quality data for fish physiology studies.
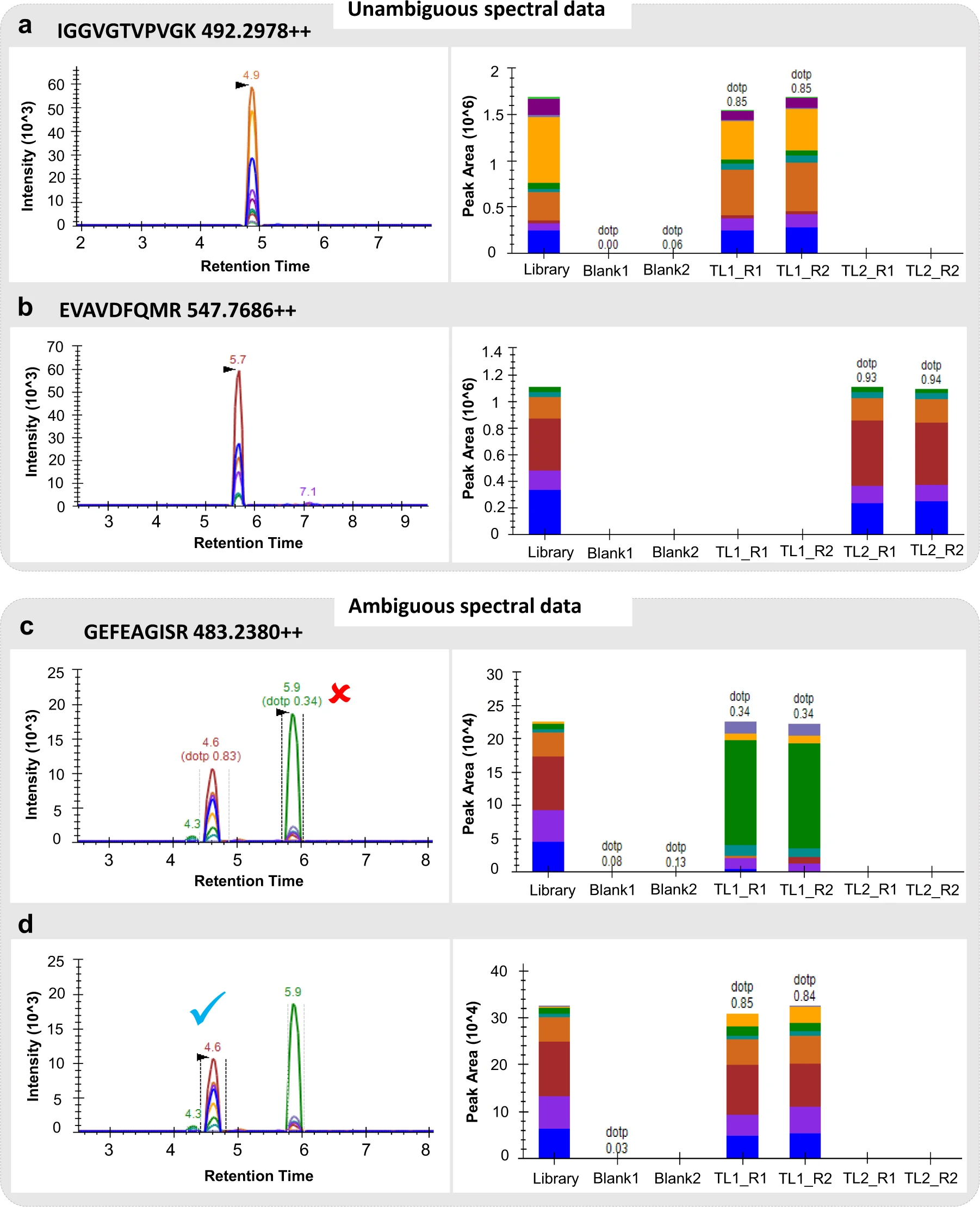
Nissa M U. et al. Scientific data. 2022.
How to order?







