Small Proteins Structure Characterization Service
Small Proteins Structure Characterization Service is a high-resolution structural analysis platform based on cryo-electron microscopy (Cryo-EM), specifically designed for proteins with molecular weights typically below 100 kDa. This service is ideal for low-molecular-weight proteins that are challenging for conventional structural techniques such as X-ray crystallography and NMR spectroscopy—particularly in cases where the protein cannot be crystallized, exhibits significant conformational flexibility, structural heterogeneity, or dynamic functional states. It enables atomic model construction without the need for crystallization and under conditions that closely mimic the native environment.
In life science research and drug discovery, small proteins play critical roles in processes such as signal transduction, enzymatic regulation, viral replication, and cellular metabolism. These proteins often possess high intrinsic flexibility, dynamic conformations, or locally disordered regions, making them difficult to crystallize or express in stable form. As a result, traditional methods like X-ray crystallography and nuclear magnetic resonance (NMR) face significant limitations when applied to these challenging targets.
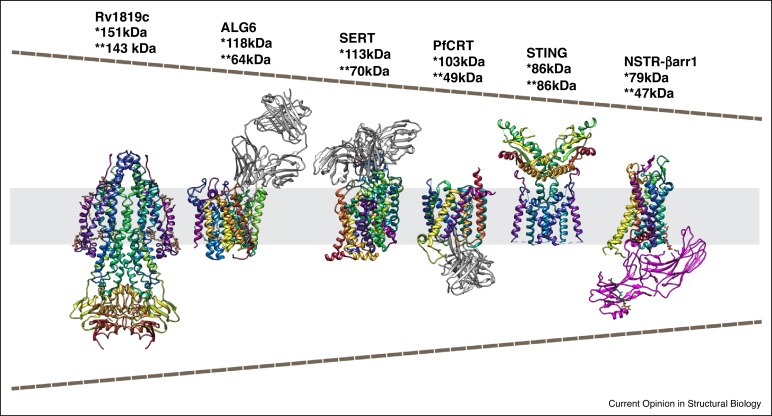
Nygaard R. et al. Curr Opin Struct Biol. 2020.
In recent years, the rapid advancement of single-particle cryo-electron microscopy (Cryo-EM SPA)—combined with next-generation direct electron detectors (e.g., Gatan K3) and powerful image processing algorithms such as TOPAZ, cryoSPARC, and RELION—has enabled Cryo-EM to overcome the traditional "molecular weight barrier," making it possible to resolve 3D structures of proteins in the 70–100 kDa range and even smaller.
Based on the advanced cryo-electron microscopy platform, MtoZ Biolabs provides Small proteins Structure Characterization Service to analyze small molecule proteins or protein complexes and output standardized atomic coordinates, conformational models and structural annotation results through highly sensitive detectors, AI particle recognition algorithms, conformational classification and labeling-assisted strategies, which is suitable for scientific research, antibody engineering, biopharmaceutical development and mechanism research.
Analysis Workflow
MtoZ Biolabs offers a comprehensive Small Proteins Structure Characterization Service covering stages from sample optimization to atomic model building:
1. Sample Preparation and Quality Assessment
Evaluate the purity, monomeric state, and conformational stability of the small protein to ensure suitability for cryo-EM analysis.
2. Grid Preparation and Negative-Stain Screening
Optimize ice thickness and particle concentration; use negative-stain TEM to assess particle distribution and sample homogeneity.
3. High-Throughput Cryo-EM Data Acquisition
Acquire large-scale 2D micrographs using cryo-EM to generate high-quality raw data for downstream 3D reconstruction.
4. 3D Reconstruction and Conformational Classification
Use particle picking, alignment, and 2D/3D classification algorithms to generate 3D density maps and identify potential conformational heterogeneity.
5. Atomic Model Building and Structural Annotation
Build atomic coordinate models of the small protein, annotate functional regions, active sites, and conformational states, and deliver a standardized structural report.
Applications
Examples of applications for the Small Proteins Structure Characterization Service include:
Structure-Function Annotation
Reveal catalytic centers, ligand-binding pockets, or conformational regulation mechanisms.
Small-Molecule Drug Target Characterization
Provide structural insights to support structure-based drug design (SBDD).
Protein Engineering and Mutation Validation
Assess structural stability, conformational changes, and the impact of key residues.
Virus-Host Interaction Studies
Analyze viral proteins such as RNA-binding proteins or non-structural proteins (NSPs), including those from SARS-CoV-2.
Structural Analysis of Key Signaling Molecules
Investigate kinases, transcription factors, E3 ligases, modifying enzymes, and other critical signaling proteins.
FAQ
Q. Can Cryo-EM Really Resolve Structures of Proteins Smaller than 100 kDa?
Yes. With the advancement of direct electron detectors (e.g., Gatan K3), phase plate technology, and AI-powered particle picking algorithms, cryo-EM now reliably resolves proteins in the 70–100 kDa range—or even smaller. The resolution can reach 3-5 Å depending on the sample characteristics.
Q. Are Additional Tags (e.g., Fab, MBP, GFP) Required for Small Proteins? Will They Affect the Native Conformation?
Yes. For proteins under 100 kDa, tags such as Fab fragments, MBP, or GFP are commonly used to enhance particle contrast, improve angular distribution, and facilitate reconstruction. However, tag design must be carefully optimized to avoid blocking functional domains or disrupting native conformations. We recommend N-terminal or flexible linker fusions, in combination with structural modeling and functional validation, to determine optimal tag placement.
Case Study
This study used cryo-electron microscopy (Cryo-EM) technology to resolve the three-dimensional structure of AmpG, a small transport protein related to bacterial cell wall metabolism. The protein has a molecular weight of approximately 58 kDa and belongs to the major facilitator family (MFS) transporter. The results showed that AmpG is composed of 14 transmembrane helices forming a unique transmembrane channel, which contains a hydrophobic vestibule structure that can recognize and transport the cell wall degradation product - 1,6-anhydro cell wall peptidoglycan (GlcNAc-anhMurNAc). Structural analysis also revealed several key substrate-binding residues (such as Lys62 and Tyr152), and their functional role in regulating β-lactam antibiotic resistance was verified through mutation experiments.
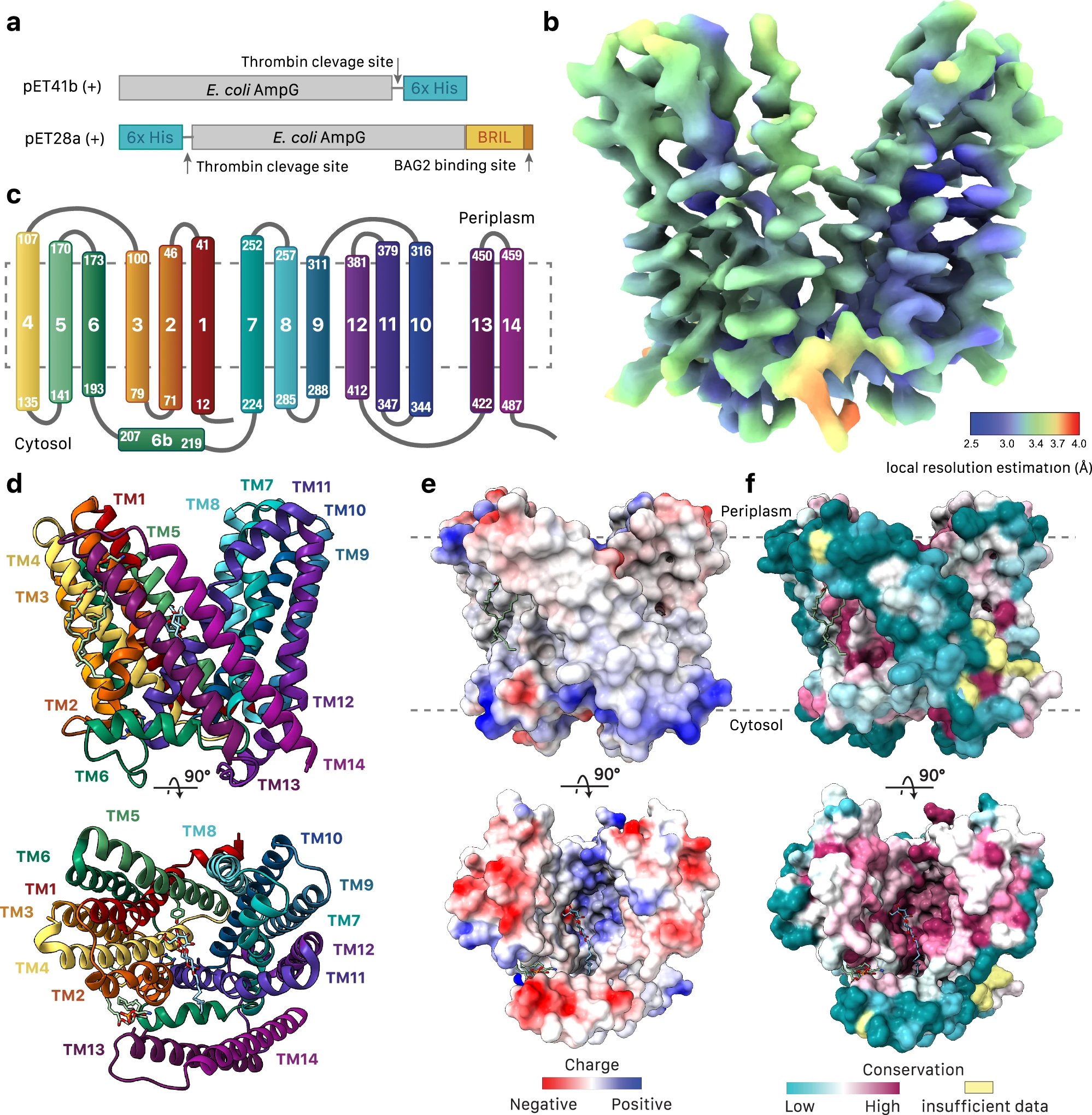
Sverak HE. et al. Nat Commun. 2024.
How to order?







