Serine/Threonine Phosphoproteomics Analysis Service
Serine/Threonine phosphoproteomics is a specialized field within proteomics that focuses on analyzing the phosphorylation modifications of serine (Ser) and threonine (Thr) residues in proteins. This approach provides critical insights into the dynamic regulatory mechanisms of cellular signaling networks. Phosphorylation, a key post-translational modification (PTM) mediated by protein kinases, governs the activation and regulation of numerous cellular processes, including metabolism, cell cycle progression, stress responses, and signal amplification. Given its dynamic and reversible nature, phosphorylation plays an essential role in biological processes, and its dysregulation is closely linked to various diseases, such as cancer, neurodegenerative disorders, immune dysfunction, and metabolic syndromes.
By leveraging advanced high-resolution mass spectrometry (MS) technology, serine/threonine phosphoproteomics enables the highly sensitive identification and quantification of phosphorylation sites, providing a comprehensive understanding of phosphorylation networks. This facilitates research on disease mechanisms, the discovery of novel biomarkers, and the identification of potential drug targets. With a state-of-the-art mass spectrometry platform and optimized phosphopeptide enrichment strategies, MtoZ Biolabs offers high-precision Serine/Threonine Phosphoproteomics Analysis Service that empowers researchers to explore phosphorylation-driven biological functions and delivering robust data to support precision medicine and drug discovery.
Technical Principles
The Serine/Threonine Phosphoproteomics Analysis Service provided by MtoZ Biolabs is based on high-resolution mass spectrometry (MS) technology, utilizing electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI) to ionize peptides for precise mass measurement and structural characterization.
Phosphorylation involves the covalent attachment of a phosphate group (H₃PO₄, ~79.98 Da), leading to an increase in protein molecular weight. When multiple phosphorylation events occur, this effect becomes cumulative. High-resolution mass spectrometry platforms, such as Orbitrap and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry (FTICR-MS), can accurately detect these subtle mass shifts. Tandem mass spectrometry (MS/MS) further pinpoints the exact phosphorylation sites. By employing data-dependent acquisition (DDA) and data-independent acquisition (DIA) strategies, Serine/Threonine Phosphoproteomics Analysis Service ensures comprehensive phosphoproteome coverage and high-precision quantification. Additionally, integrating results with expert-curated databases (e.g., UniProt, PhosphoSitePlus) enables systematic identification of phosphorylated proteins and their functional roles in signaling pathways.
Analysis Workflow
To ensure high coverage and precision, our Serine/Threonine Phosphoproteomics Analysis Service follows a streamlined, systematic workflow:
1. Sample Preparation: Collection of cell, tissue, or biofluid samples, followed by lysis, protein extraction, and trypsin digestion to generate peptide mixtures.
2. Phosphopeptide Enrichment: Selective enrichment of low-abundance phosphopeptides using IMAC or TiO₂ to enhance detection sensitivity.
3. LC-MS/MS Analysis: High-resolution LC-MS/MS platforms (e.g., Thermo Fisher Orbitrap series) are used for phosphopeptide separation and identification. DDA or DIA strategies enable precise quantitative analysis.
4. Data Analysis & Bioinformatics Interpretation: Phosphorylation sites are identified using curated databases (e.g., UniProt, PhosphoSitePlus). Downstream analyses include pathway enrichment, kinase-substrate prediction, and dynamic phosphorylation profiling to extract key biological insights.

Service Advantages
MtoZ Biolabs' Serine/Threonine Phosphoproteomics Analysis Service offers several key advantages:
· High-Sensitivity Detection: Optimized phosphopeptide enrichment strategies allow for precise detection of low-abundance phosphorylation events.
· Comprehensive Phosphoproteome Coverage: Advanced LC-MS/MS technology ensures extensive proteomic depth.
· Accurate Quantitative Analysis: Integration of TMT, DIA, or label-free techniques enables precise relative and absolute quantification.
· Expert Bioinformatics Interpretation: In-depth phosphorylation site identification, pathway enrichment analysis, and kinase-substrate prediction provide valuable functional insights.
Sample Submission Suggestions
To achieve optimal experimental outcomes, we recommend submitting the following biological sample types, ensuring they meet quality standards for mass spectrometry analysis:
For specialized sample needs, MtoZ Biolabs offers tailored experimental solutions and sample processing optimization services.
Applications
The Serine/Threonine Phosphoproteomics Analysis Service supports a wide range of research and industrial applications, including but not limited to:
· Cell Signaling Pathway Studies: Identifying key proteins involved in cellular responses by analyzing phosphorylation patterns.
· Disease Research: Investigating disease-associated phosphorylation changes to identify potential biomarkers.
· Drug Discovery: Evaluating how candidate drugs modulate protein phosphorylation.
Case Study
Case 1
Using mass spectrometry, this study identified multiple phosphorylation sites on the NIPA protein mediated by NPM-ALK, including Ser-338, Ser-344, Ser-370, Ser-381, and Thr-387. These phosphorylation events are crucial for NIPA localization and its interaction with NPM-ALK, ultimately influencing cellular proliferation. The study demonstrated the Serine/Threonine Phosphoproteomics Analysis Service as a valuable tool for disease mechanism research, particularly in analyzing protein phosphorylation dynamics and identifying disease-related therapeutic targets.
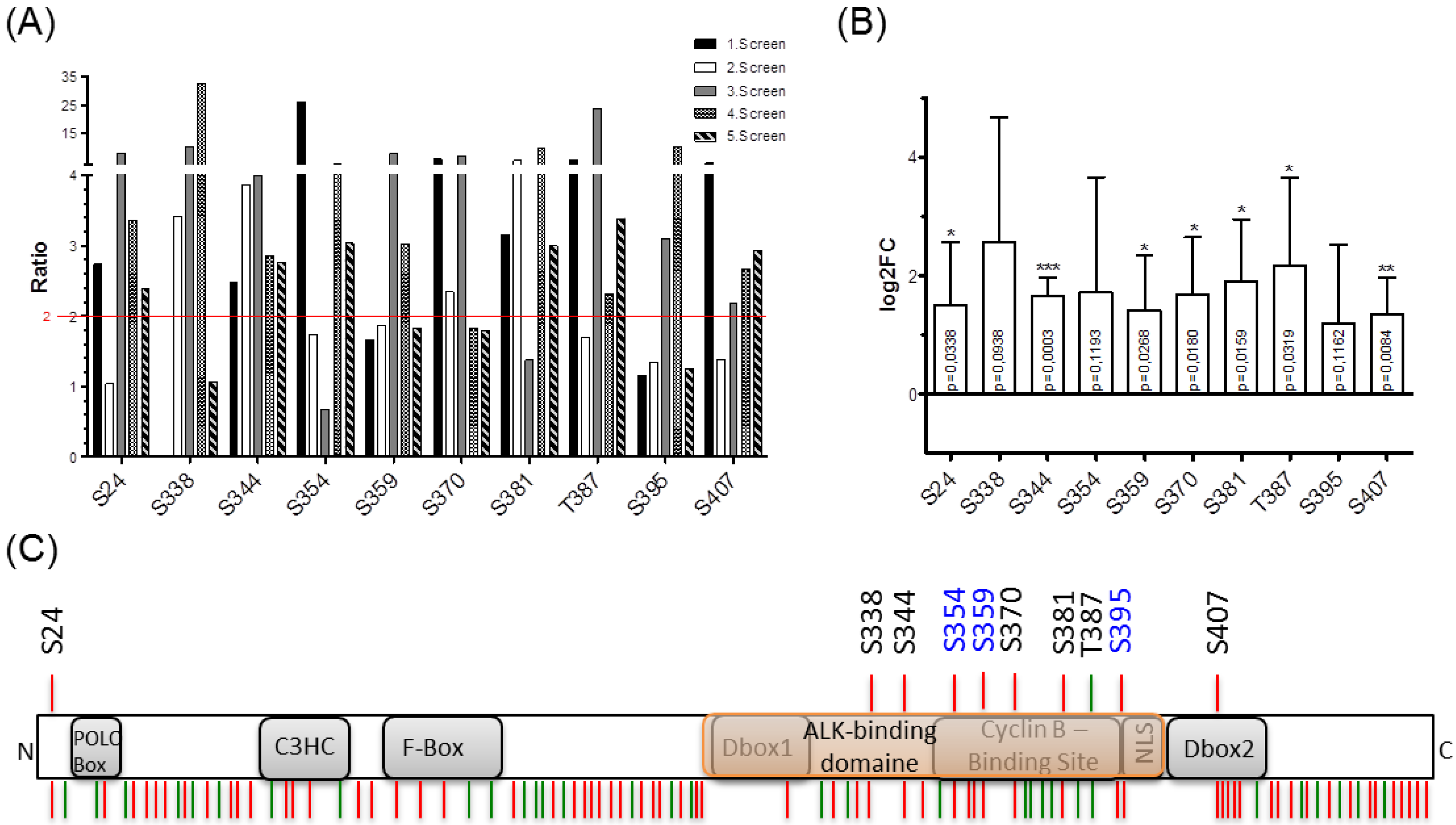
Gengenbacher, A. et al. Int. J. Mol. Sci. 2019.
Case 2
This study employed proteomics and phosphoproteomics approaches to investigate the roles of serine/threonine kinase (STK) and phosphatase (STP) in Streptococcus suis, particularly in bacterial regulatory mechanisms. The results showed that STK plays a more significant role than STP in cell division, energy metabolism, and pathogenicity. Phenotypic analysis of mutant strains and phosphorylation profiling further confirmed these findings. This study highlights the Serine/Threonine Phosphoproteomics Analysis Service as an effective approach for elucidating bacterial regulatory networks.
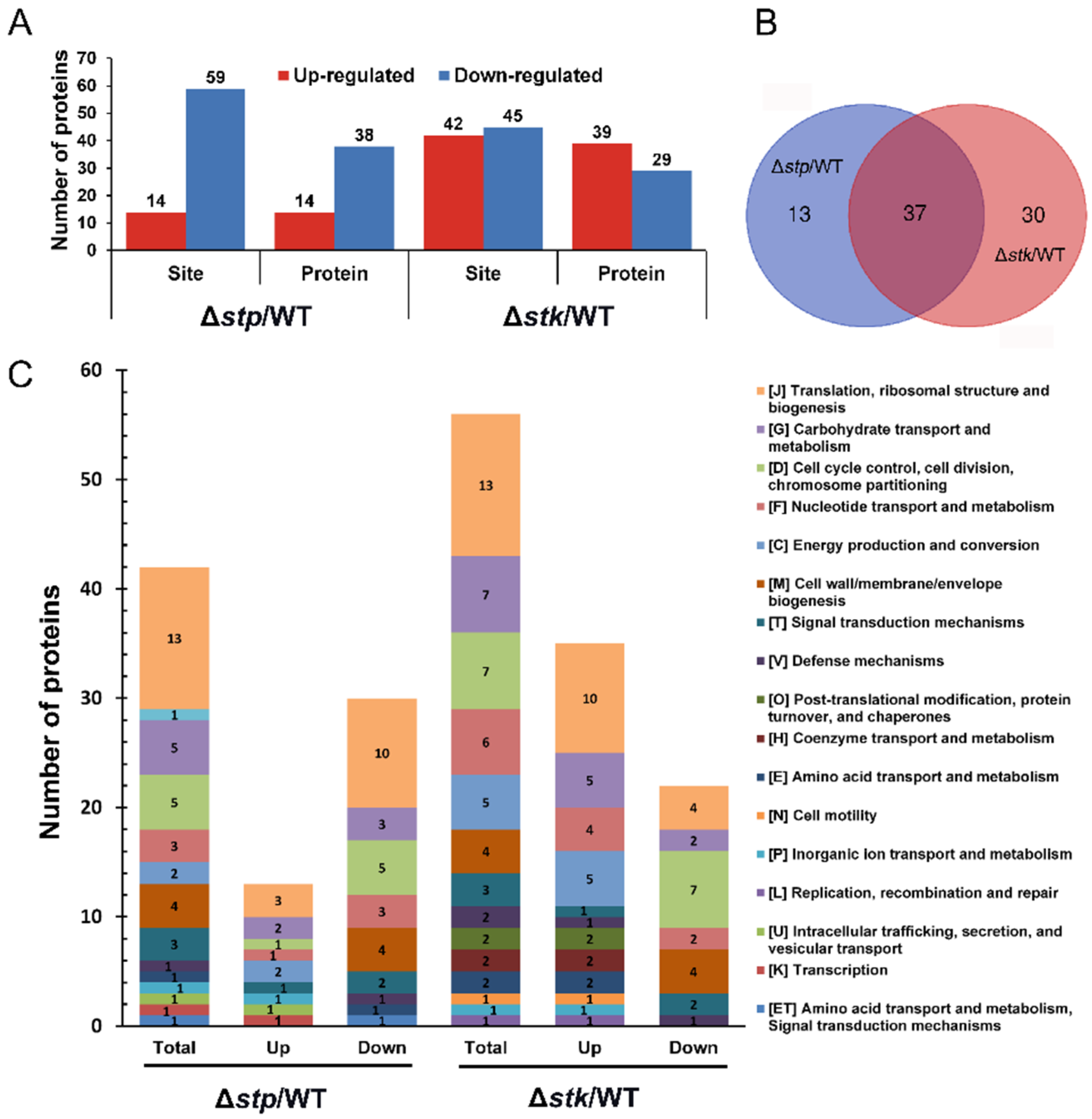
Hu, Q. et al. Microorganisms. 2021.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
With its cutting-edge mass spectrometry platform, optimized phosphopeptide enrichment techniques, and expert bioinformatics team, MtoZ Biolabs delivers high-sensitivity, high-coverage Serine/Threonine Phosphoproteomics Analysis Service. If you have research needs in phosphorylation analysis, feel free to contact us for professional consultation and customized experimental solutions.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Phosphoproteomics Service
How to order?







