S-Glutathionylation Proteomics Service
S-Glutathionylation Proteomics is an advanced strategy for systematically investigating the functional roles of S-glutathionylation, a reversible cysteine modification critical for redox regulation. This approach integrates high-resolution mass spectrometry, selective enrichment, and bioinformatics-driven annotation to reveal how this modification influences protein function, antioxidant defense, and disease-related signaling pathways.
S-glutathionylation involves the covalent attachment of glutathione (GSH) to protein thiols, forming mixed disulfide bonds that protect key enzymatic sites and maintain redox homeostasis. It is increasingly recognized as a regulatory mechanism and potential biomarker in oxidative stress-associated conditions such as cardiovascular disease, cancer, and neurodegenerative disorders.
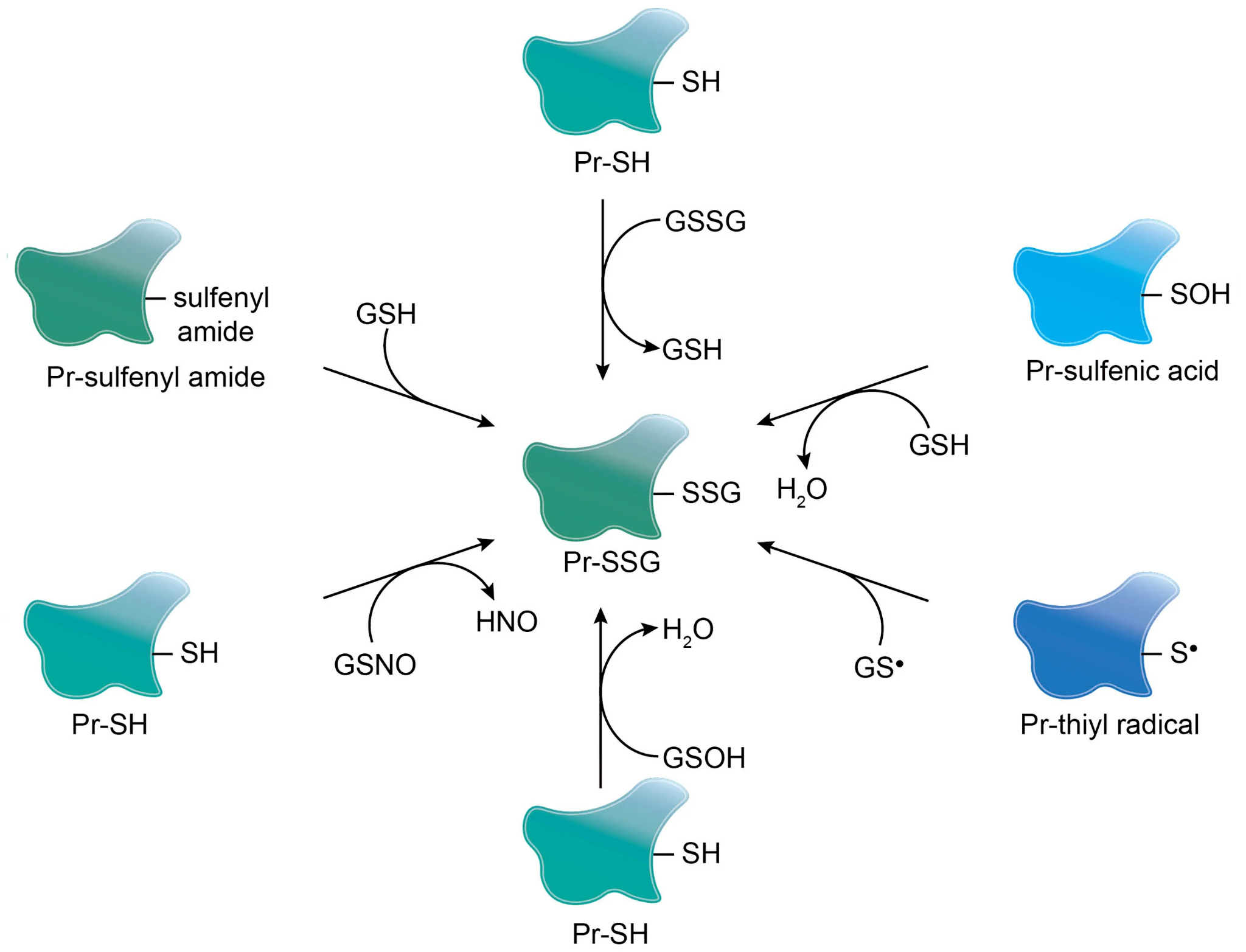
Kalinina, E. V. et al. Biochemistry Moscow. 2023.
Figure 1. Mechanisms of Spontaneous S-Glutathionylation
Leveraging advanced instrumentation and optimized workflows, MtoZ Biolabs offers a dedicated S-Glutathionylation Proteomics Service to support in-depth analysis of modification sites and their biological relevance across physiological and pathological contexts.
Services at MtoZ Biolabs
To address the low abundance and dynamic nature of S-glutathionylation modifications, our S-Glutathionylation Proteomics Service offers a highly sensitive and high-throughput solution for modification site identification and functional interpretation, including:
· Selective Peptide Enrichment
Affinity capture using anti-GSH antibodies or thiol-reactive resins improves coverage and specificity of SSG-modified peptides.
· High-Resolution Site Identification
Cysteine-centered PTM mapping with sub-ppm mass accuracy on Orbitrap platforms (Fusion, Lumos, Q Exactive HF).
· Quantitative Profiling
Label-Free or TMT-based quantification enables comparative analysis across treatment groups or time courses.
· Structure-Function Correlation
Use of predictive modeling tools to visualize SSG-modified sites in the context of protein domains or enzymatic cores.
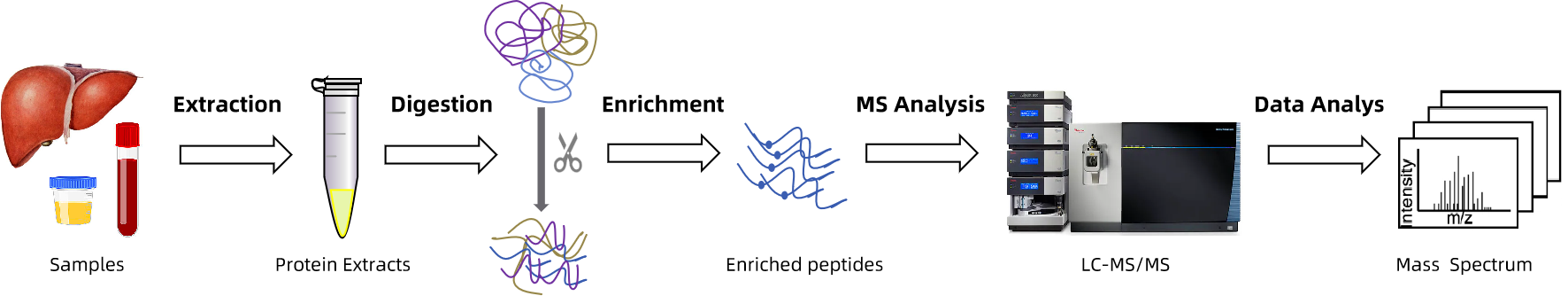
Analysis Workflow
Our S-glutathionylation workflow combines optimized sample preparation with Nano-LC–Orbitrap MS acquisition and downstream analysis using established PTM pipelines. The platform has been successfully applied to phosphorylation, glycosylation, ubiquitination, and disulfide bond analysis—ensuring both sensitivity and modification specificity.
Why Choose MtoZ Biolabs?
✅ High-resolution MS capability: Sub-ppm mass accuracy and deep proteome coverage enabled by Orbitrap Fusion Lumos and Q Exactive HF platforms
✅ Optimized enrichment workflow: GSH-targeted capture strategy enhances signal-to-noise ratio and recovers low-abundance S-glutathionylation sites
✅ Multi-PTM analysis compatibility: Supports co-enrichment and analysis with phosphorylation, ubiquitination, and nitrosylation for PTM crosstalk mapping
✅ End-to-end project adaptability: Flexible options for sample preparation, experimental design, and data reporting tailored to research needs
✅ Broad sample compatibility: Applicable to cell lysates, tissues, plasma/serum, and purified proteins across various biological systems
Applications
Our S-Glutathionylation Proteomics Service supports multiple research and preclinical use cases:
· Redox and stress signaling: Characterize dynamic redox-sensitive cysteine modifications under oxidative challenge
· Disease mechanism exploration: Construct SSG modification maps in models of cardiovascular, oncological, or neurodegenerative diseases
· Drug response and antioxidant evaluation: Track SSG shifts in response to GSH modulators, redox-targeted therapies, or dietary interventions
· Environmental toxicology: Study S-glutathionylation changes in response to pollutants, radiation, or chemical exposures
· Biomarker discovery: Identify site-specific SSG events with diagnostic or therapeutic potential
Case Study
Case 1: Mapping S-glutathionylation on Cardiac Myosin via Top-Down Proteomics
In a study, researchers applied top-down mass spectrometry to map endogenous S-glutathionylation (SSG) on ventricular myosin light chain-1 (MLC-1v). The team detected native SSG-modified forms in human and pig hearts—but not in mice—highlighting species-specific redox regulation.
By supplementing non-reduced lysates with oxidized glutathione (GSSG), they confirmed the inducibility of the modification. Quantitative analysis revealed significant SSG elevation across all species. These findings underscored S-glutathionylation’s regulatory impact on contractile proteins and cardiac redox balance.
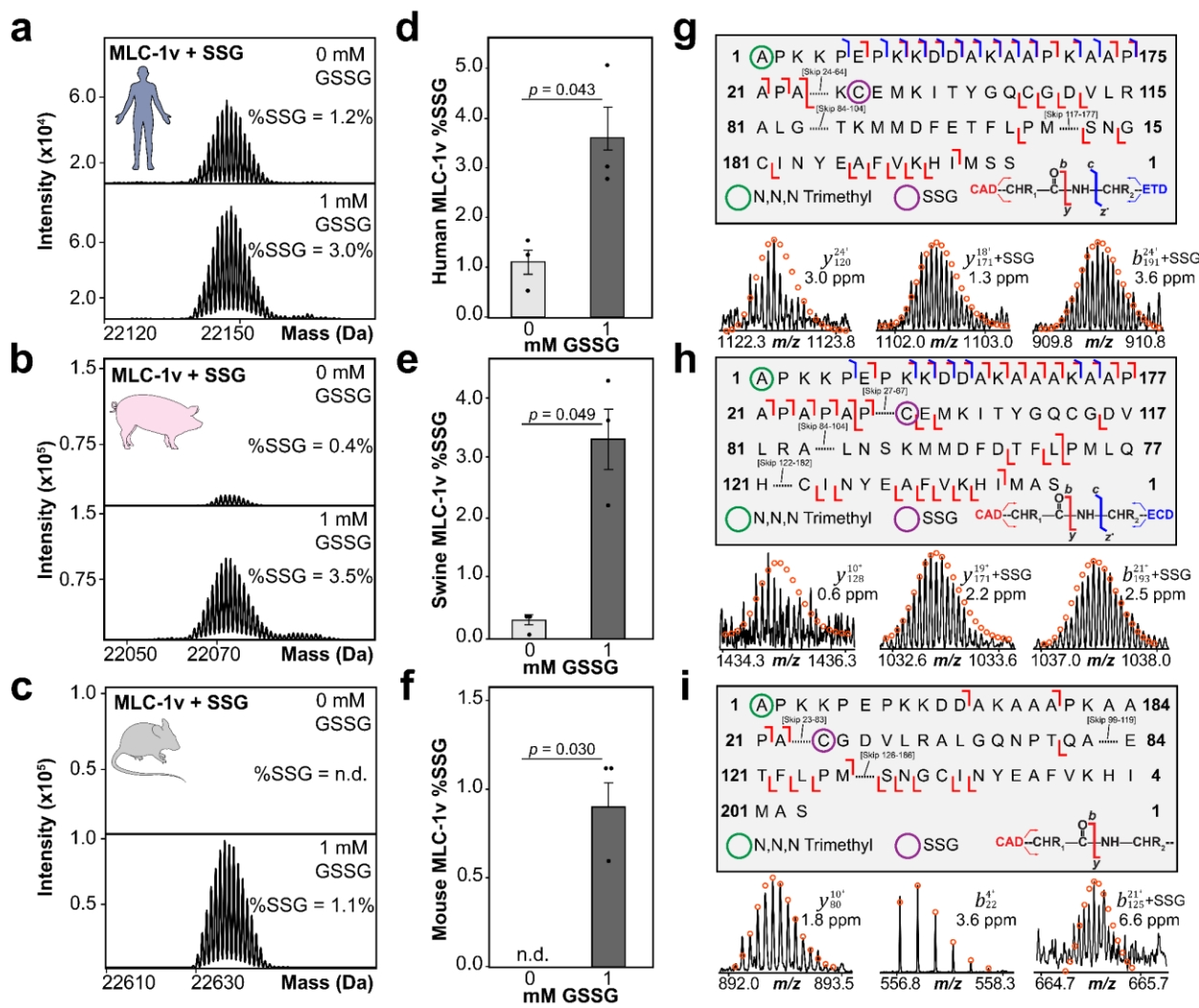
Chapman, E. A. et al. J MOL CELL CARDIOL. 2025.
MtoZ Biolabs’ S-Glutathionylation Proteomics Service enables similar high-sensitivity SSG detection and quantification—ideal for redox, cardiovascular, and translational proteomics research.
FAQ
Q1: How can I avoid loss or reversal of S-glutathionylation during sample prep?
We recommend blocking free thiols with alkylating agents like NEM or IAM during protein extraction to stabilize the modification. All samples should be snap-frozen and shipped under cold-chain conditions to preserve in vivo SSG states.
Q2: Can SSG data be integrated with other PTM datasets?
Yes. We offer integrated multi-PTM analysis, allowing SSG to be studied alongside phosphorylation, ubiquitination, and more—ideal for modeling crosstalk networks in oxidative stress or signal transduction.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
At MtoZ Biolabs, we combine expertise in proteomics, redox biology, and PTM research to offer unmatched support for S-glutathionylation analysis. Our S-Glutathionylation Proteomics Service empowers you to define oxidative modification patterns, understand their regulatory mechanisms, and translate findings into therapeutic insights.
Contact us today for project consultation and customized analysis solutions.
How to order?







