Proteomics Bioinformatics Analysis Service
Proteomics Bioinformatics Analysis is the process of processing, analyzing, and visualizing proteomic data obtained through mass spectrometry using computational methods and statistical tools. Its primary goal is to extract valuable insights from large-scale proteomics datasets to reveal protein expression patterns, interactions, functional annotations, and biological significance. This analysis relies on database searches (e.g., UniProt, PDB), algorithm modeling (e.g., machine learning, network analysis), data normalization, and statistical analysis to ensure precise protein identification and quantification. Common methods include Data-Dependent Acquisition (DDA) and Data-Independent Acquisition (DIA) data analysis, differential protein analysis, functional enrichment analysis (GO/KEGG), protein-protein interaction (PPI) network construction, and multi-omics integration.
Proteomics bioinformatics analysis service has extensive applications across various fields, including disease mechanism research, biomarker discovery, precision medicine, drug target screening, immunology research, and agricultural and food sciences. For example, in disease research, this analysis can be used to identify key proteins and regulatory pathways involved in cancer and neurodegenerative diseases. In drug development, it enables the evaluation of candidate drugs' effects on the proteome, assessing their mechanisms of action and safety. Additionally, proteomics bioinformatics can be applied to post-translational modification (PTM) studies, investigating cellular signaling pathways and metabolic regulation, providing strong support for life sciences research.
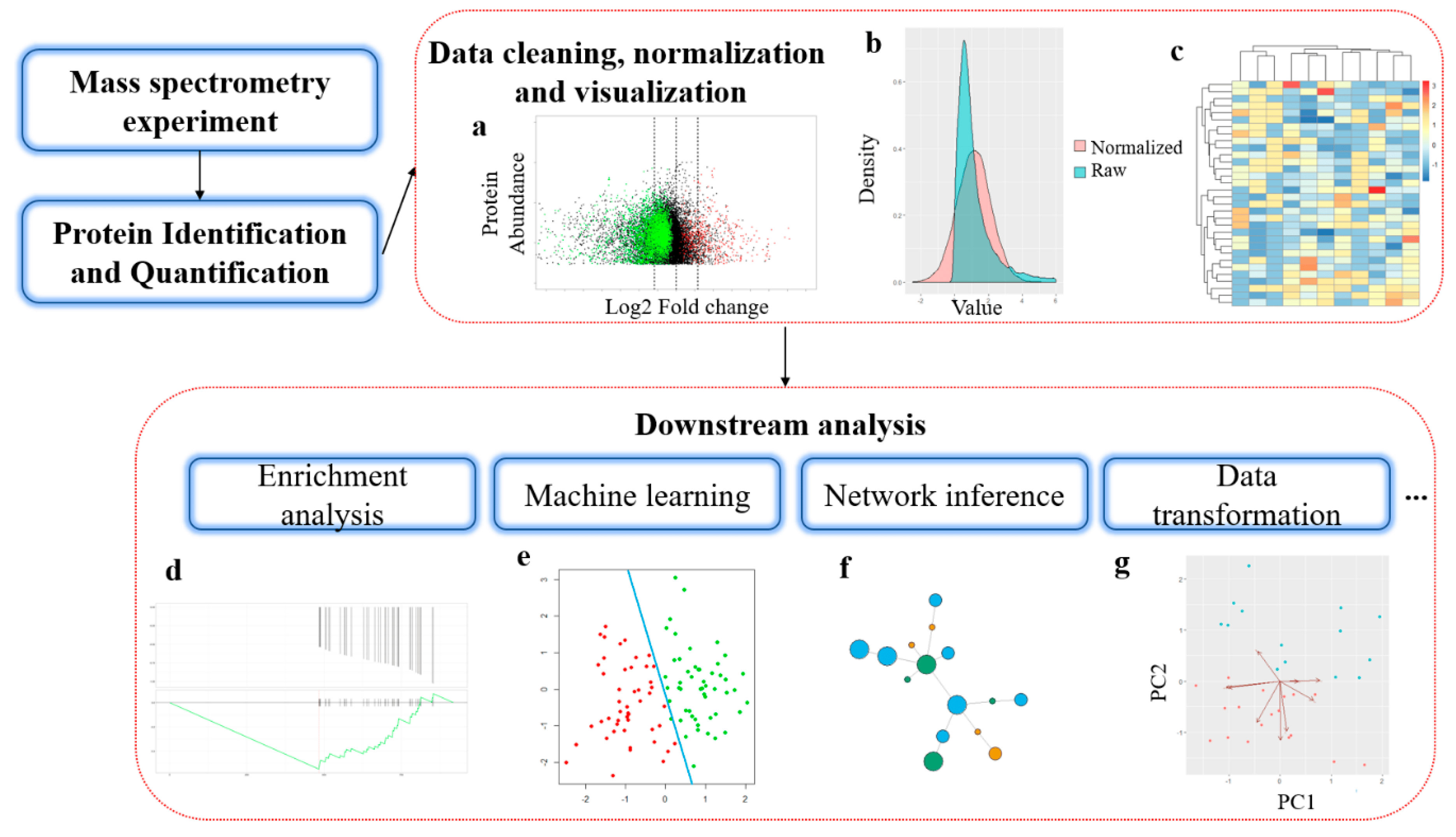
Chen, C. et al. International Journal of Molecular Sciences, 2020.
Figure 1. General Workflow of Bioinformatics Analysis in Mass Spectrometry-based Proteomics.
Services at MtoZ Biolabs
Based on advanced high-resolution mass spectrometry and computational platforms, MtoZ Biolabs offers proteomics bioinformatics analysis service to ensure precise processing and in-depth interpretation of proteomics data. We provide a comprehensive analysis workflow, covering data preprocessing, database searching, quantitative analysis, and biological functional annotation. Our services support various mass spectrometry data types, including DIA, DDA, and PRM. The main aspects of our services include:
1. Proteomic Data Quality
2. Differential Protein Statistical Analysis
3. GO Functional Annotation and Enrichment Analysis
4. KEGG Pathway and Enrichment Analysis
5. COG Functional Annotation and Enrichment Analysis
6. Differentially Expressed Proteins Custer Analysis
7. Protein Interaction Analysis
Service Advantages
1. High-Precision Data Processing
MtoZ Biolabs integrates advanced algorithms and computational platforms to offer proteomics bioinformatics analysis service that provide in-depth analysis of DIA, DDA, and PRM mass spectrometry data. This ensures highly accurate protein identification and reliable quantitative data.
2. Comprehensive Biological Functional Annotation
We offer differential protein analysis, GO/KEGG pathway enrichment, and protein-protein interaction (PPI) network construction, enabling researchers to uncover the biological functions and regulatory mechanisms of proteins.
3. Multi-Omics Integration Analysis
MtoZ Biolabs supports the integration of proteomics with transcriptomics, metabolomics, and epigenomics, constructing systems biology networks to enhance the depth and breadth of research.
4. Customized Analytical Solutions
We provide tailored solutions for specific signaling pathway analysis, post-translational modification (PTM) studies, and biomarker discovery, catering to diverse applications in basic research, disease studies, and drug development.
Applications
1. Disease Research and Biomarker Discovery
Through quantitative proteomics analysis, we identify differentially expressed proteins in healthy and diseased states, facilitating the discovery of potential biomarkers. This supports the early diagnosis and precision treatment of major diseases such as cancer, neurodegenerative disorders, and cardiovascular diseases.
2. Drug Target Identification and Mechanism of Action Studies
Proteomics bioinformatics analysis service enables the identification of drug targets, investigation of drug mechanisms, and assessment of drug-induced changes in protein expression and post-translational modifications. These insights contribute to new drug development and personalized treatment strategies.
3. Cell Signaling Pathways and Functional Analysis
By integrating GO/KEGG enrichment analysis and protein-protein interaction (PPI) network construction, proteomics bioinformatics analysis service helps elucidate protein functions in cell signaling, metabolic regulation, and immune responses.
4. Multi-Omics Integration Analysis
By combining proteomics with transcriptomics and metabolomics, our approach enables a deeper understanding of biological regulatory mechanisms. This supports precision medicine, biopharmaceutical development, and biotechnological research.
Case Study
1. Mining cancer biology through bioinformatic analysis of proteomic data
This study aims to explore cancer-related biological mechanisms through bioinformatics analysis of proteomic data and to identify potential cancer biomarkers and therapeutic targets. The study utilizes proteomic datasets from various cancer types, obtained from public databases or clinical samples collected through experimental methods. High-resolution mass spectrometry (LC-MS/MS) was employed to acquire protein expression profiles, which were subsequently analyzed using bioinformatics tools such as GO annotation, KEGG pathway analysis, protein-protein interaction (PPI) network construction, and differential expression analysis. The findings reveal that specific key proteins and their post-translational modifications (PTMs) play crucial roles in signaling pathways, metabolic regulation, cell cycle control, and immune evasion across different cancer types. Additionally, multiple potential cancer biomarkers and therapeutic targets were identified, with validation confirming their potential involvement in cancer progression. The study concludes that proteomics bioinformatics analysis provides novel insights into cancer research, facilitating the investigation of cancer pathogenesis, the identification of precision therapy targets, and the advancement of personalized medicine.
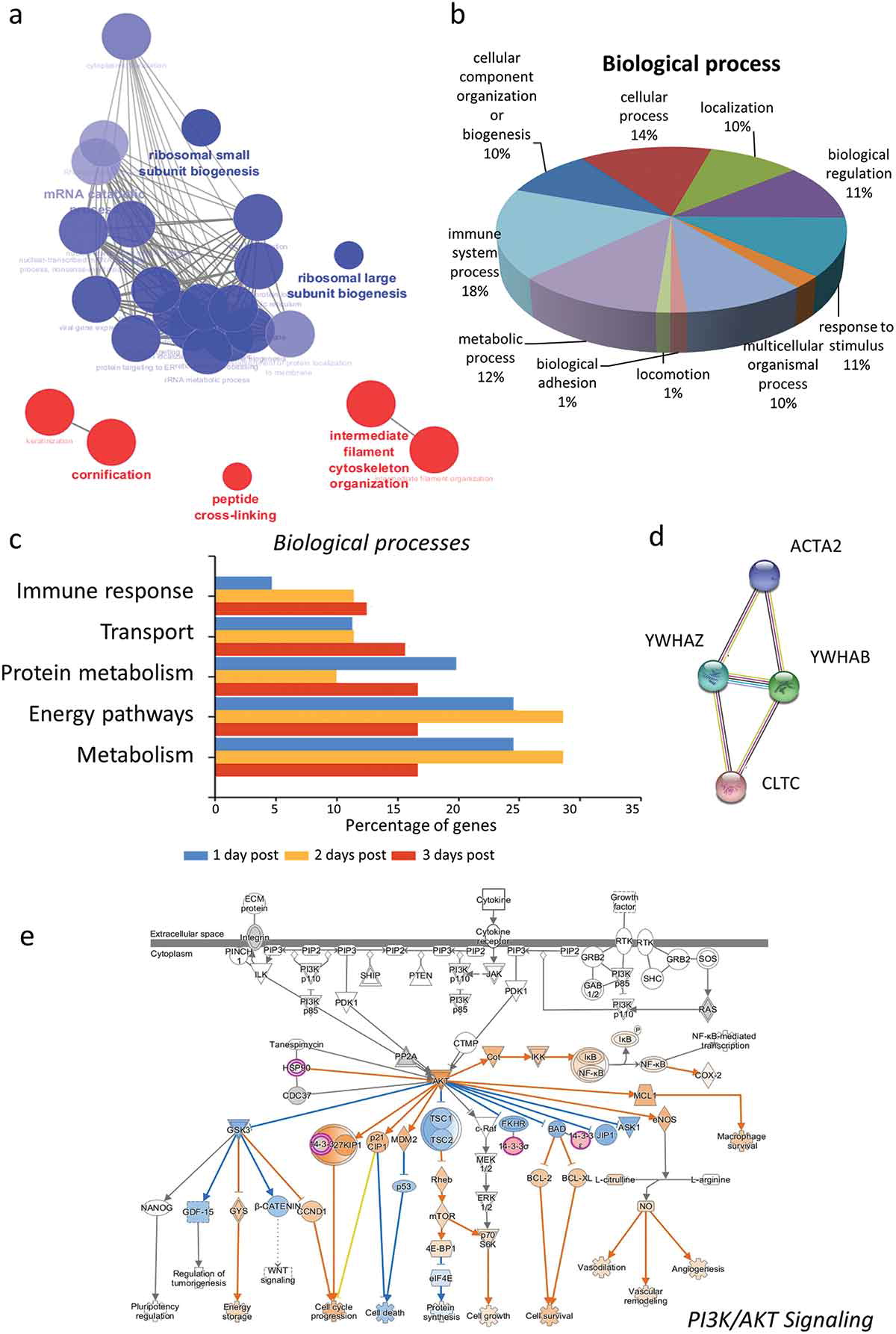
Manfredi, M. et al. Expert Review of Proteomics, 2019.
Figure 2. Different Types of Data Representation in Bioinformatics.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
How to order?







