Protein Quantification Service
Protein quantification measures the expression levels of proteins under different conditions to reveal their functions and dynamic regulation in biological processes, serving as a critical tool for studying biomarkers, molecular mechanisms, and drug targets. Relative quantification compares protein abundance between samples, while absolute quantification provides specific concentration information. Protein quantification is essential for identifying potential diagnostic and therapeutic biomarkers, elucidating key roles in signaling pathways and metabolic networks, and evaluating drug target functionality. It delivers high-resolution and wide dynamic range data, enabling researchers to better understand the complexity of biological systems and providing significant scientific insights for disease research, drug development, and precision medicine.
Service at MtoZ Biolabs
MtoZ Biolabs offers advanced protein quantification services that cater to the diverse needs of both research and industry sectors for precise protein analysis. For relative quantification, the company utilizes highly sensitive mass spectrometry technologies and efficient protein labeling methods to effectively compare changes in protein expression levels across different samples, aiding researchers in uncovering regulatory mechanisms and potential biomarkers in biological processes. In absolute quantification, the service employs standards with known concentrations and optimized analytical workflows to accurately determine the absolute amounts of target proteins in samples, which is crucial for disease diagnosis, drug development, and quality control of biological products. With a professional technical team and state-of-the-art equipment, MtoZ Biolabs is dedicated to providing reliable data, advancing life sciences research, and facilitating the successful implementation and innovative breakthroughs of scientific projects.
1. Relative Protein Quantification
Here describes a protocol for relative quantification using TMT 8plex reagents. The TMT 8plex technique is used to describe the analysis procedure, including protein digestion, TMT reaction, sample purification, fractionation, HPLC-MS, and data analysis. Eight different protein samples are denatured, reduced, alkylated and digested into peptide samples. These peptide samples are then labeled with 8plex TMT reagent and then combined into one sample. The combined sample is fractionated using HPRP liquid chromatography and the fractions are concatenated into 10 samples. After cleaning up, each sample is subjected to HPLC-MS analysis and the data are then analyzed to obtain the identity and relative quantification of proteins.
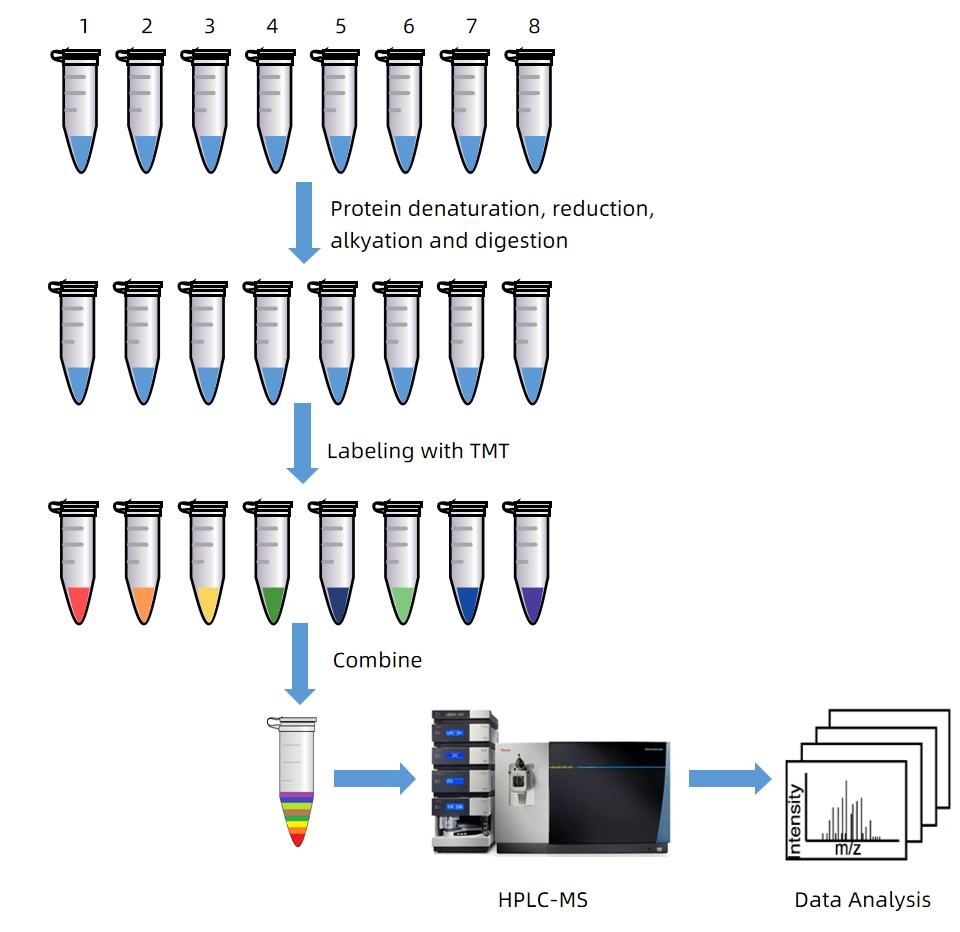
Figure 1. Workflows of Relative Protein Quantification Based on 8plex TMT Labeling
2. Absolute Protein Quantification
Absolute protein quantification refers to the process of accurately determining the absolute amount of a target protein or peptide in a sample, such as its mass, molar concentration, or copy number. This approach typically involves sample extraction and purification, preparation or addition of standards, separation and detection of the target substance, and final quantitative analysis. Mass spectrometry techniques (such as ESI-MS or ICP-MS) combined with internal standards, labeling methods, or standard curves are key to achieving absolute quantification. By comparing with standards of known concentrations, absolute quantification provides precise evaluations of the true protein content in complex samples.
Nosti, AJ. et al. J Proteomics. 2022.
Figure 2. Workflows of Top-down and Bottom-up Absolute Quantitative Proteomics Combining Elemental and Molecular Mass Spectrometry
Service Advantages
1. Comprehensive and Optimized Protein Extraction
MtoZ Biolabs employs a variety of efficient extraction methods to ensure complete cell lysis and high protein recovery. These methods are adaptable to different organisms and sample types, minimizing protein loss and ensuring accurate quantification.
2. Reliable Absolute Quantification Methods
MtoZ Biolabs offers multiple absolute quantification strategies, utilizing stable isotope standards and calibration curves to accurately convert mass spectrometry signals into protein concentrations. This ensures data accuracy and supports critical applications.
3. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
4. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all protein quantification data, providing clients with a comprehensive data report.
Applications
1. Relative Protein Quantification in Preclinical Drug Discovery
Schirle, M. et al. Chem Biol. 2012.
2. Absolute Protein Quantification for Research into Disease Biomarker
Kumar, A. et al. Sci Rep. 2017.
FAQ
Question 1: When performing TMT protein quantification, why is SDS-PAGE used first?
Answer: SDS-PAGE is typically used as a preliminary step for several reasons:
1. Sample Quality Assessment
SDS-PAGE can separate and display the protein components in a sample, allowing for a visual evaluation of sample quality through the electrophoresis pattern. This helps in checking the following aspects:
(1) Protein Concentration: Ensuring that the protein concentration in the sample is appropriate for TMT labeling and mass spectrometry analysis.
(2) Protein Integrity: Assessing whether proteins are degraded or denatured to ensure high-quality samples.
(3) Protein Composition: Observing the presence of expected proteins in the sample and detects any unexpected contaminants or impurities.
2. Confirmation of Sample Homogeneity
Before TMT labeling, SDS-PAGE can help confirm that the protein composition across different samples is consistent. If significant differences are observed, further processing or optimization of sample preparation steps may be necessary to ensure the accuracy and comparability of TMT quantification results.
3. Selection of Appropriate Protein Digestion Points
By determining the size and distribution of proteins through SDS-PAGE, suitable cut-off points for enzymatic digestion (such as trypsin digestion) can be selected. This optimization enhances subsequent mass spectrometry analysis by improving peptide recovery and detection sensitivity.
4. Removal of Interfering Substances
SDS-PAGE can aid in removing substances that might interfere with TMT labeling and mass spectrometry analysis, such as salts, lipids, and other small molecule impurities. By excising specific protein bands after electrophoresis, purer samples can be obtained.
5. Verification of Protein Labeling Efficiency
Separating labeled proteins using SDS-PAGE allows for the assessment of TMT labeling efficiency and specificity. This ensures that the labeling reaction is complete and uniform, thereby enhancing the accuracy and reliability of TMT quantification.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of Protein Quantification
5. Mass Spectrometry Image
6. Raw Data
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Proteomics Service
How to order?







