Protein Molecular Weight Determination by Gel Filtration: Key Points, and Practical Applications
Protein molecular weight determination plays a crucial role in understanding the structure, function, and biological properties of proteins. Gel Filtration Chromatography (GFC), also known as Size Exclusion Chromatography (SEC), is a technique that separates proteins based on molecular size. This method is extensively used in protein purification, molecular weight estimation, and complex formation analysis. This paper delves into the foundational principles, essential experimental considerations, data interpretation, and real-world applications of protein molecular weight determination by gel filtration.
Principles of Gel Filtration
1. Mechanism of Action
Gel filtration chromatography employs porous beads as stationary phase to separate proteins by size. As the protein sample traverses the column, smaller proteins penetrate the pores, while larger proteins are excluded and elute first.
(1) Large proteins cannot enter the pores and elute quickly, resulting in a smaller elution volume (Ve).
(2) Small proteins can enter the pores, leading to delayed elution and a larger Ve.
This size-based separation enables gel filtration to be utilized for both purification and molecular weight determination of proteins.
2. Key Parameters
(1) Volume Parameters
①Total Column Volume (Vt): Total volume of the column
②Void Volume (Vo): Elution volume for molecules completely excluded from the pores (often measured with large markers like blue dextran)
③Elution Volume (Ve): The volume at which the target protein is eluted
④Relative Elution Volume (Kav): Calculated to estimate protein molecular weight
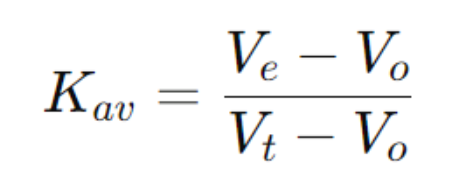
Figure 1
(2) Gel Media
①Typically composed of materials such as dextran (Sephadex), agarose (Sepharose), polyacrylamide (Bio-Gel), or silica gel.
②The selection of appropriate pore sizes is critical for effective separation of the target protein.
Key Points in Protein Molecular Weight Determination by Gel Filtration
1. Choosing Appropriate Gel Media
(1) Fractionation Range: Should align with the target protein's molecular weight, e.g., Sephadex G-75 (3-70 kDa) or Superdex 200 (10-600 kDa).
(2) If the protein's molecular weight significantly deviates from the media's range, separation efficiency diminishes.
2. Calibration Curve
A series of standard proteins with known molecular weights is used for calibration to plot the standard curve:
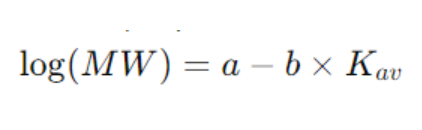
Figure 2
Where MW represents the molecular weight, and Kav is determined experimentally. The molecular weight of an unknown protein is estimated using the standard curve.
3. Sample Preparation
(1) Ensure thorough dissolution without precipitates to prevent column clogging.
(2) Filter or centrifuge to remove particulates.
(3) Add DTT or β-mercaptoethanol to break disulfide bonds and avoid aggregation.
4. Experimental Conditions
(1) Flow Rate: Maintain a low flow rate (0.2-1 mL/min) to preserve resolution.
(2) Buffer: Should match protein properties to prevent denaturation, using buffers like PBS, Tris-HCl, or Hepes.
(3) Temperature: Conduct at low temperatures (e.g., 4°C) to minimize protein degradation.
5. Data Analysis
(1) Record the elution peaks (UV 280 nm) to determine Ve.
(2) Use the standard curve to calculate the protein's molecular weight.
(3) Multiple peaks may indicate dimers, aggregates, or complexes.
Practical Applications of Protein Molecular Weight Determination by Gel Filtration
1. Estimating Native Protein Molecular Weight
Gel filtration assesses the molecular weight of proteins in their native state, unlike SDS-PAGE, which indicates the denatured state. For example, a monomer at 30 kDa on SDS-PAGE might appear as a 60 kDa peak, suggesting dimerization.
2. Analyzing Protein Complexes
Evaluate protein interactions with other molecules. For instance:
(1) Increased molecular weight in nucleic acid-binding proteins suggests binding with DNA/RNA.
(2) Changes in immune complex size indicate antigen-antibody interactions.
3. Detecting Protein Purity and Aggregates
Essential in biopharmaceuticals to detect aggregates that may affect function or immunogenicity.
4. Monitoring Conformational Changes
Gel filtration can track changes in proteins under varying pH, temperature, or ion concentrations. For example, chaperone proteins like Hsp70 may form different oligomeric states in response to ATP.
Advantages and Limitations of Gel Filtration
1. Advantages
(1) Mild Separation: Preserves native structure, ideal for determining molecular weight of native proteins.
(2) High Resolution: Differentiates aggregation states such as monomers, dimers, or tetramers.
(3) Macromolecular Complex Analysis: Useful for studying interactions like protein-protein or protein-nucleic acid.
(4) Versatile with HPLC/FPLC: Enhances separation efficiency.
2. Limitations
(1) Lower Accuracy than Mass Spectrometry: Provides approximate molecular weights without precise sequence details.
(2) High Sample Stability Required: Proteins must remain stable to avoid aggregation or precipitation.
(3) Limited Resolution of Similar Sizes: Ineffective for proteins with close molecular weights.
Gel Filtration Chromatography (SEC) is pivotal in applications such as protein molecular weight determination, complex analysis, aggregate detection, and conformational studies. Despite its lower precision compared to mass spectrometry, its non-denaturing approach is invaluable for studying native protein states. Advances in media and automation, like high-resolution SEC-HPLC and multi-angle light scattering (MALS), will further enhance its role in biomedicine, proteomics, and structural biology. MtoZ Biolabs is dedicated to providing high-quality protein molecular weight determination services, utilizing cutting-edge gel filtration methods for accurate measurements, offering reliable data for research. For more information on the key points and applications of gel filtration methods, we welcome collaboration to advance scientific progress.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Protein Molecular Weight Determination Service
Gel Filtration Chromatography Molecular Weight Determination Service
How to order?







