Protein Characterization Service
Protein characterization service refers to the comprehensive analysis of the physicochemical properties, molecular structure, post-translational modifications (PTMs), and molecular interactions of proteins, among others. Proteins serve as the carriers of life substances and executors of biological functions, playing a crucial role in maintaining normal physiological activities of organisms. As one of the core areas in biological science research, protein characterization analysis reveals how proteins function within biological systems and how they interact to maintain normal physiological functions. MtoZ Biolabs provides efficient and precise protein characterization services to drive scientific innovation and industrial applications.
Common methods for protein characterization analysis include nuclear magnetic resonance (NMR), mass spectrometry (MS), and Western blotting (WB). NMR is commonly used to study the three-dimensional structure and dynamic properties of proteins. MS analyze the molecular weight, post-translational modifications, and interactions of proteins. While WB utilizes specific antibodies to detect distances and interactions between proteins.
Services at MtoZ Biolabs
MtoZ Biolabs' protein characterization service integrates these technological platforms, leveraging advanced instruments such as Orbitrap mass spectrometers and Cryo-Electron Microscopy (Cryo-EM), to deliver comprehensive solutions from molecular weight determination to structural analysis and functional validation. This service is widely applied in drug development, biomarker discovery, and fundamental life science research, providing researchers with high-resolution and reliable data support.
1. Protein Purity Analysis
Protein characterization service includes the assessment of protein purity using various methods such as SDS-PAGE, capillary electrophoresis, zinc reverse staining, glutaraldehyde-modified silver staining, and MALDI-TOF-MS. These techniques are employed to detect contaminants, protein variants, isoforms, S-S bond mismatches, truncated proteins, degraded proteins, protein modifications, protein aggregates, and protein precursors.
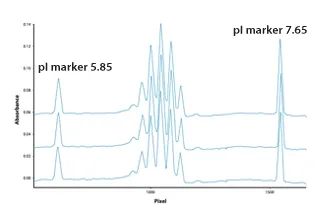
Figure 1. Characterization of Protein Purity and Uniformity Marked by pI
2. Protein Molecular Weight Analysis
Protein complexes and protein-small molecule complex molecular weights are determined using techniques such as gel permeation chromatography, SDS-PAGE, and mass spectrometry.
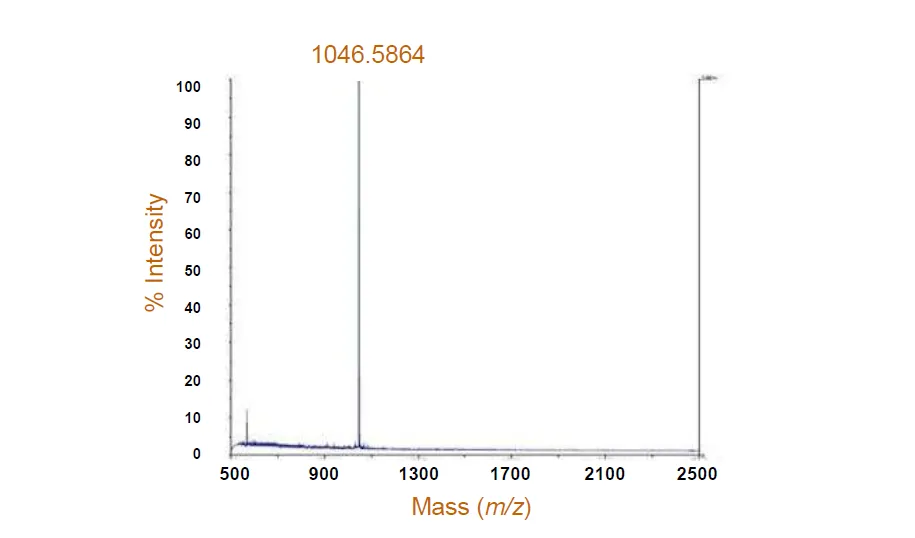
Figure 2. Molecular Weight Determination
3. Protein Structure Identification
Protein characterization service provides precise structural insights, including top-down primary structure characterization (such as amino acid composition analysis, peptide sequence analysis, and disulfide bond localization) using Thermo Fisher's Orbitrap Fusion Lumos mass spectrometry platform combined with nanoLC-MS/MS. Additionally, spatial configuration determination is achieved using circular dichroism spectroscopy.
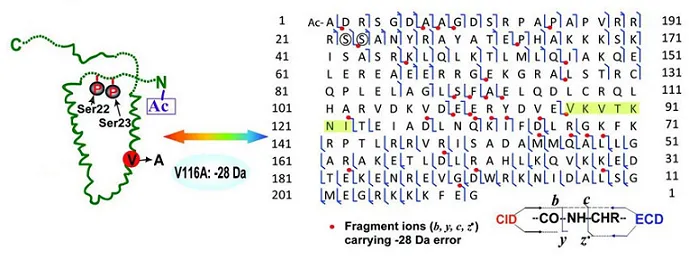
Figure 3. Identification of Protein Primary Structure
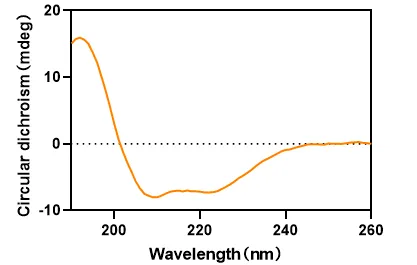
Figure 4. Samples' CD Spectra in the Far-Ultraviolet Range (190-260 nm)
4. Protein Post-Translational Modification Analysis
Multiple mass spectrometry platforms (Thermo Fisher's Q Exactive HF, Orbitrap Fusion, and Orbitrap Fusion Lumos mass spectrometry platforms combined with Nano-LC) are employed for the identification of protein phosphorylation, glycosylation, ubiquitination, acetylation, methylation, disulfide bond formation, nitrosylation, and among other post-translational modifications.
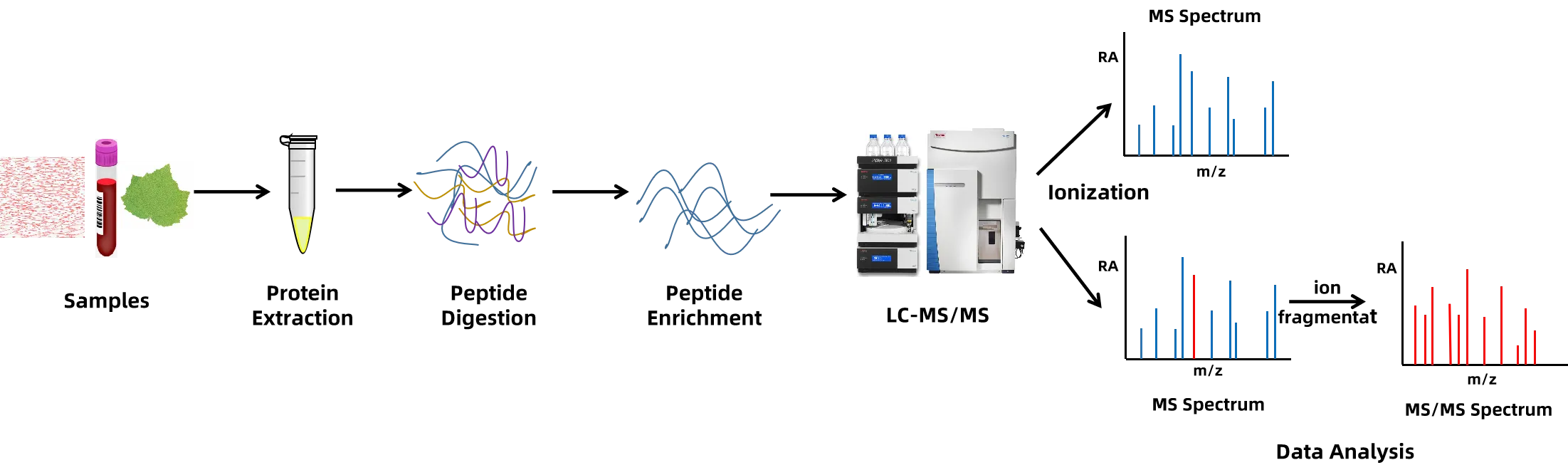
Figure 5. Analysis Workflow of Protein Post-Translational Modification
5. Protein Interaction Analysis and Identification of Interacting Proteins
Techniques such as WB, immunoprecipitation (IP), and GST-pull-down are utilized to study protein-protein interactions. LC-MS/MS is employed for mass spectrometric identification analysis of proteins/protein complexes in IP, Co-IP samples, and GST fusion protein pull-down purified samples.
Service Advantages
1. Accurate Analysis of Complex Product Formats
Our protein characterization service employs advanced LC-MS technologies, including native LC-MS and multidimensional LC-MS methods, enabling comprehensive characterization of complex biotherapeutic formats while preserving higher-order structures for precise attribute analysis.
2. Adaptability to Diverse Biotherapeutic Needs
Protein characterization service in MtoZ Biolabs is tailored to various types of protein-based therapeutics, such as IgG-like molecules and derivatives, by optimizing separation techniques and detection methods to address the increasing complexity of formats.
3. Integration of Multidimensional Data Analysis
Protein characterization service provided by MtoZ Biolabs integrates two-dimensional or multidimensional LC-MS applications, combined with UV/FLD detection, to comprehensively evaluate key product attributes, delivering in-depth structural and functional insights.
Applications
1. Protein Characterization Service for Biophysical Characterization of Antibody-Based Therapeutics
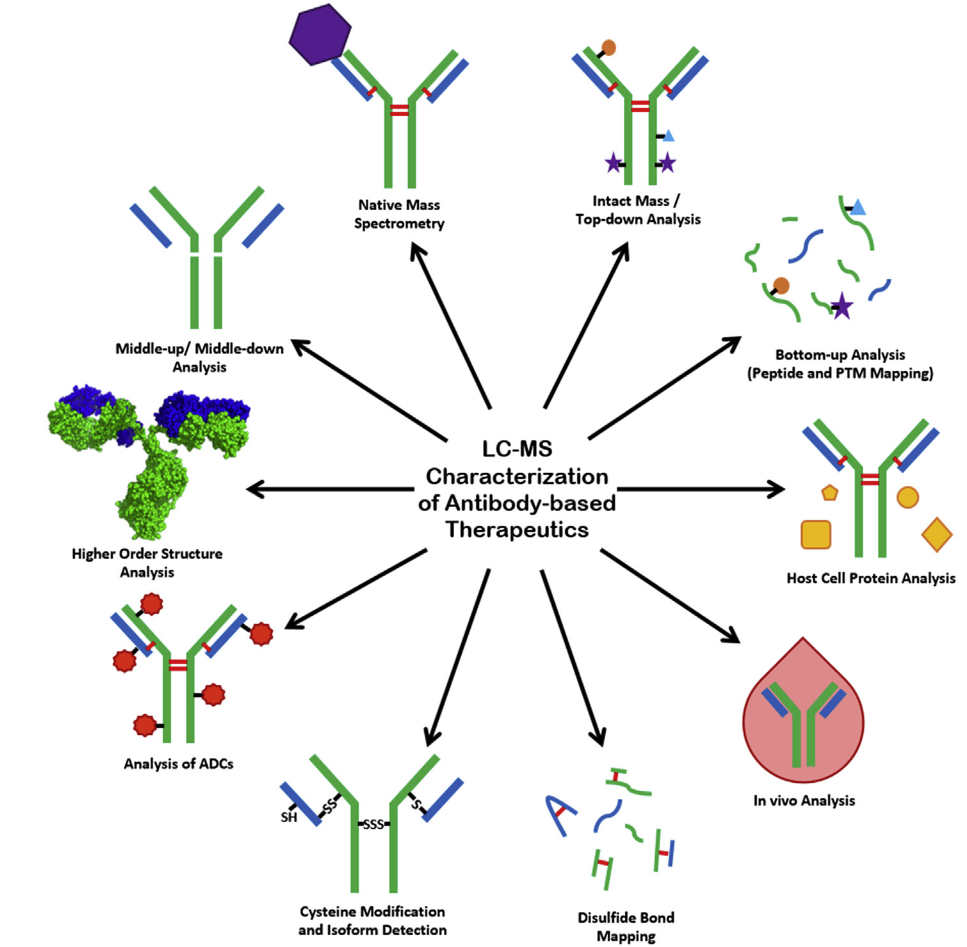
Anna, C. et al. 2020.
Figure 6. Protein Characterization Service for Biophysical Characterization of Antibody-Based Therapeutics
Deliverables
1. Experimental Procedures
2. Relevant Mass Spectrometry Parameters
3. Detailed information on protein characterization analysis
4. Mass Spectrometry Images
5. Raw Data
MtoZ Biolabs' Protein Characterization Service is designed to provide comprehensive and high-quality solutions for researchers and industry professionals. From analyzing protein purity, molecular weight, and structural properties to exploring post-translational modifications and protein interactions, our advanced platforms and specialized expertise ensure precise, reliable, and reproducible results. By leveraging cutting-edge technologies such as Thermo Fisher's Orbitrap mass spectrometers, Nano-LC systems, and a range of supplementary methodologies, we address even the most challenging protein characterization needs. Contact us now to discuss how we can support your success!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Protein Structure Identification Service
How to order?







