Principles of Olink Proteomics
In high-throughput proteomics research, a long-standing challenge has been achieving sensitive and specific protein detection from minimal sample volumes. The Olink® proteomics platform addresses this by introducing the Proximity Extension Assay (PEA), which ingeniously combines the specificity of antibody recognition with the high sensitivity of nucleic acid amplification, thereby overcoming fundamental limitations of conventional protein detection techniques.
Core Mechanism of Olink Proteomics: Proximity Extension Assay (PEA)
1. Dual-Antibody Recognition Mechanism
Each target protein is recognized by a pair of high-affinity antibodies, each conjugated to a distinct oligonucleotide probe (DNA sequence). These probes are brought into close spatial proximity only when both antibodies simultaneously bind to the same protein molecule. This proximity-dependent event functions as a dual-recognition safeguard, significantly enhancing detection specificity and minimizing interference from non-specific binding.
2. Proximity-Dependent DNA Extension
Once the DNA probes are co-localized on the target protein, they undergo a proximity extension reaction that forms a unique DNA sequence. This sequence serves as a molecular identifier for the specific protein and acts as a template for subsequent nucleic acid amplification. The term “Proximity Extension Assay” reflects this process: “Proximity” refers to spatial closeness, and “Extension” denotes the formation of a new DNA product.
3. QPCR or NGS-Based Amplification and Detection
The resulting DNA template can be amplified and quantified using two approaches:
(1) Target Panel (qPCR-based detection): Real-time quantitative PCR (RT-qPCR) performed in 96- or 384-well plates, suitable for small-scale and multi-sample studies.
(2) Explore platform (NGS-based detection): Library preparation followed by sequencing via Illumina high-throughput platforms, ideal for ultra-high-throughput needs (detecting 3000+ proteins).
This exponential nucleic acid amplification enables the detection of proteins at extremely low concentrations (fg/mL), particularly benefiting the measurement of low-abundance proteins such as cytokines, hormones, and inflammatory mediators.
Data Output: How Does NPX Reflect Protein Abundance?
The final output from the Olink platform is the NPX (Normalized Protein Expression) value, a log-transformed expression value derived from normalized Ct values (qPCR) or read counts (NGS).
Key features of NPX include:
(1) Unit-independent relative expression values.
(2) Suitable for intergroup comparison, clustering analysis, and heatmap visualization.
(3) While not representing absolute concentration, NPX offers excellent dynamic range and batch-to-batch consistency, making it suitable for cross-sectional studies.
MtoZ Biolabs offers advanced NPX-based data mining services including differential expression analysis, pathway enrichment, and machine learning modeling to support comprehensive interpretation of Olink datasets.
Summary of Technical Advantages of Olink Proteomics
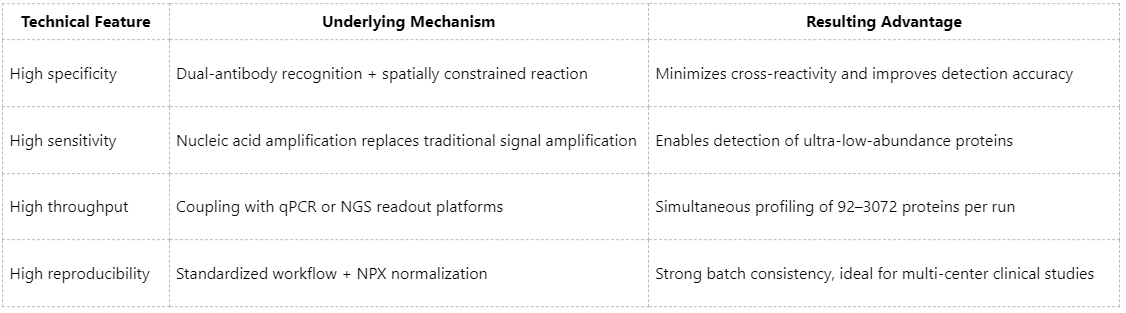
Core Applications of Olink Proteomics
The PEA technology at the heart of Olink proteomics facilitates a range of cutting-edge applications, including:
1. Multi-omics integration: Joint modeling of proteomic, transcriptomic, and metabolomic data to uncover novel regulatory pathways.
2. Early disease prediction: Identification of early biomarkers at the subclinical stage enabled by ultra-sensitive detection.
3. Drug response monitoring: Longitudinal profiling of protein expression dynamics before and after therapeutic intervention.
4. Interstitial fluid/CSF analysis: Effective profiling of ultra-low-abundance proteins that are challenging for traditional mass spectrometry.
MtoZ Biolabs has successfully executed multiple research and clinical projects using the Olink platform, offering end-to-end support including: standardized guidance for sample preprocessing (especially low-abundance samples); assistance in selecting optimal Panel combinations; full project management aligned with Olink-certified workflows; and expert bioinformatics support for differential analysis, network modeling, and biological annotation. The widespread adoption of the Olink proteomics platform is driven by its innovative PEA technology, which synergistically combines dual-antibody specificity, proximity-driven DNA extension, and nucleic acid-based detection to deliver breakthroughs in sensitivity, specificity, throughput, and reproducibility in protein analysis.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







