Preparing Samples for Protein Sequencing
-
Supports a wide range of sample types, including tissues, cells, body fluids, and exosomes
-
Employs automated processing platforms to enable high-throughput preparation without batch effects
-
Provides pre-loading peptide quality control reports to ensure data integrity
-
Technical team can tailor customized solutions for specific experimental goals (e.g., phosphorylation, post-translational modifications, or labeled quantification)
In proteomics, mass spectrometry (MS) serves as the central analytical technique, with its results heavily dependent on sample quality. Regardless of the analytical approach—whether comprehensive proteome analysis, modification-focused studies, or targeted protein quantification—high-quality sample preparation is a prerequisite. This paper systematically examines the sample preparation workflow for protein sequencing, encompassing pretreatment strategies for various sample types, protein extraction, enzymatic digestion, purification, and quality control standards. Additionally, it introduces MtoZ Biolabs' solutions for each of these steps.
Sample Types and Challenges Prior to Protein Sequencing
1. Common Sample Sources
(1) Tissue samples: including liver, brain, and tumor tissues, commonly used in clinical research or animal models
(2) Cell samples: cultured or primary cells, offering advantages in experimental control
(3) Body fluid samples: such as serum, plasma, urine, and cerebrospinal fluid, well-suited for biomarker discovery
(4) Trace samples: including exosomes and single cells, which present challenges due to low total protein content
2. Challenges in Sample Preparation
(1) Wide variations in protein concentration and dynamic range
(2) Interference from lipids, DNA, and salts, which can hinder enzymatic digestion and instrument performance
(3) Protein denaturation, aggregation, or degradation, leading to analytical bias
Standard Protein Sequencing Sample Preparation Workflow
1. Cell/Tissue Lysis
Lysis of samples is the initial step for total protein extraction and requires consideration of both lysis efficiency and protein stability. Commonly used lysis buffers include:
(1) RIPA buffer: suitable for most cell types and compatible with phosphoprotein analysis
(2) Urea/thiourea lysis buffer: effective for tissue samples, providing strong denaturation and facilitating membrane protein solubilization
(3) SDS buffer: applied for SDS-PAGE analysis or Filter-Aided Sample Preparation (FASP)
It is recommended to include protease and phosphatase inhibitors to prevent protein degradation and loss of post-translational modifications.
2. Protein Quantification and Quality Control
Accurate quantification forms the basis for standardized sample processing. The commonly employed methods are as follows:
(1) BCA method (recommended): This method exhibits high resistance to interference and is compatible with most lysis buffers.
(2) Bradford method: This method is characterized by operational simplicity but exhibits sensitivity to certain types of buffer systems.
(3) SDS-PAGE preliminary analysis: This method is used to assess protein integrity and to determine the extent of degradation.
3. Protein Reduction and Alkylation
Ensure complete unfolding of proteins and exposure of peptide bonds to facilitate subsequent enzymatic digestion.
(1) Reducing agents: DTT or TCEP, used to disrupt disulfide bonds
(2) Alkylating agents: IAA or CAA, used to prevent the reformation of disulfide bonds
(3) Incubation at 37°C for 30–60 minutes is optimal for reaction efficiency
4. Enzymatic Digestion (Trypsin Digestion)
Trypsin is widely recognized as the reference enzyme for proteolytic digestion prior to mass spectrometry. The recommended digestion ratio is 1:50–1:100 (enzyme:substrate), typically with overnight incubation at 37°C.
(1) Additional proteases such as Lys-C or Glu-C may be used to enhance proteome coverage
(2) Desalting or dilution is advisable prior to digestion to prevent inhibition of enzymatic activity by high concentrations of urea
5. Desalting and Purification
Following digestion, residual salts, impurities, and remaining trypsin must be removed to improve mass spectrometric sensitivity.
(1) C18 solid-phase extraction (SPE) columns: retain peptides while eliminating low-molecular-weight impurities
(2) StageTips method: an optimal approach for handling trace samples
(3) Desalting criteria: reduce sample conductivity to below 1 μS/cm
6. Pre-loading Quality Control
Sample quality must be evaluated prior to mass spectrometric analysis:
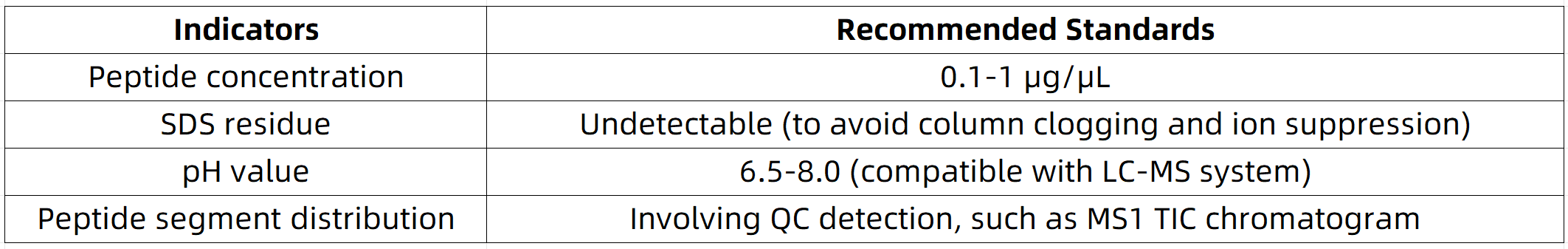
Specific requirements for sample preparation for different applications:

MtoZ Biolabs Protein Sequencing Service
At MtoZ Biolabs, we have established a standardized, traceable, and mass spectrometry-compatible sample preparation workflow encompassing every step from sample lysis, enzymatic digestion, and desalting to quality control:
Whether for fundamental research or clinical projects, we are committed to supporting your proteomics research with professional and efficient sample processing. High-quality sample preparation forms the foundation of reliable proteomics data. Only by ensuring standardized, controlled, and mass spectrometry-compatible preparation processes can the accuracy and reliability of downstream data be guaranteed. MtoZ Biolabs is dedicated to providing professional sample processing support to help your proteomics research take a confident first step.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







