Phosphoproteomics Solutions
- Snap-freezing: Instant enzyme deactivation by liquid nitrogen freezing.
- Inhibitor Application: Adding Sodium fluoride (NaF) in lysis buffer to inhibit phosphatase activity.
- Low-temperature Handling: Performing all steps at 4°C to prevent enzyme reactivation.
- Extremely Low Abundance: Phosphorylated proteins account for less than 0.1% of total proteins.
- Ionization Suppression: Phosphate groups reduce peptide ionization efficiency.
- Signal Interference: High-abundance non-modified peptides can interfere with detection.
The phosphoproteomics solutions integrate high-resolution mass spectrometry with advanced enrichment strategies to systematically analyze the dynamic changes in protein phosphorylation modifications. This approach uncovers the regulatory mechanisms of key signaling pathways, providing precise molecular insights for cell signaling, disease research, and drug target discovery. Phosphorylation is one of the most widely studied post-translational modifications (PTMs), influencing cellular signaling, metabolic regulation, cell cycle, differentiation, and the onset and progression of various diseases. Studies show that over 30% of human proteome proteins undergo dynamic phosphorylation, and abnormalities in these modification sites are closely linked to more than 200 pathological processes, including cancer and neurodegenerative diseases.
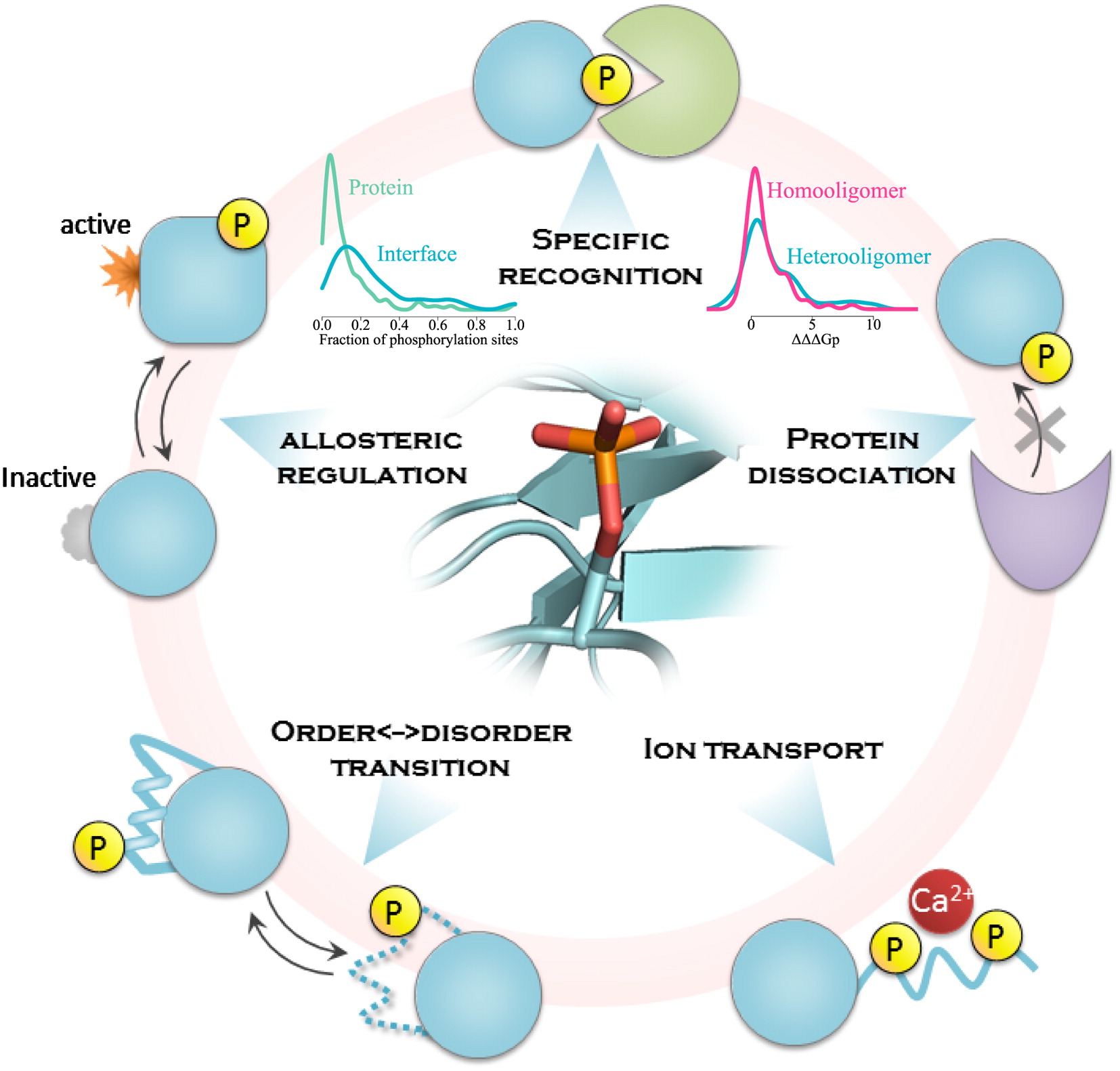
Nishi, H. et al. Structure. 2011.
Figure1. Phosphorylation in Cellular Signal Regulation: Mechanisms and Challenges
Through high-throughput and precise phosphoproteomics research, scientists are able to deeply explore the dynamic changes in signaling pathways, uncover the molecular mechanisms of diseases, and provide solid data support for the discovery of novel drug targets. However, phosphoproteomics research faces three major technical challenges:
1. Capturing low-abundance phosphopeptides (phosphorylated proteins often represent less than 0.1% of the proteome)
2. The transient nature and spatial heterogeneity of dynamic modifications
3. Precise identification of functional sites in vast amounts of data
Service at MtoZ Biolabs
As an innovative platform focused on post-translational modification proteomics, MtoZ Biolabs provides end-to-end phosphoproteomics solutions through integrated high-sensitivity mass spectrometry, phosphorylation enrichment strategies, and AI-driven bioinformatics analysis, delivering:
✅ Precise phosphorylation site identification
✅ Dynamic modification quantification
✅ Kinase-substrate network modeling
Our services support disease mechanism research, targeted drug development, and optimization of precision medicine strategies. We employ an advanced high-resolution mass spectrometry (HRMS) platform, coupled with Nano-LC separation technology and phosphorylation enrichment strategies (such as TiO₂ and IMAC), to achieve highly sensitive detection of low-abundance phosphorylation modifications. Additionally, our bioinformatics analysis platform allows for deep mining of large-scale data, helping researchers decode complex phosphorylation signaling networks and accelerate biomedical research. Our technological framework has been successfully applied in cutting-edge fields such as tumor targeted therapy and kinase inhibitor development, with detection sensitivity reaching the amol level and site identification accuracy exceeding 98%.
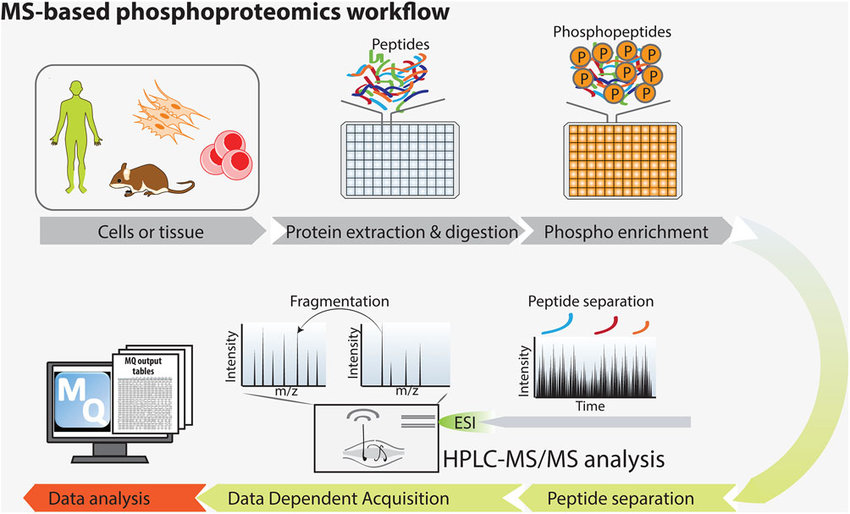
Sacco, F. et al. Proteomics. 2018.
Figure2. MS-based Phosphoproteomics Workflow
We provide comprehensive phosphoproteomics solutions, from experimental design to data analysis, including but not limited to:
1. Global Phosphoproteomics Analysis
Utilizing IMAC/TiO₂ enrichment, we enhance the detection capability of low-abundance phosphorylated peptides. This approach is ideal for global phospho-modification identification, revealing complex cellular signaling pathways.
2. Quantitative Phosphoproteomics
Based on TMT/iTRAQ or Label-free quantification strategies, we precisely measure changes in phosphoproteins under different conditions. This is suitable for drug mechanism studies and monitoring dynamic changes in signaling pathways.
3. Kinase Target Analysis
Through specific substrate screening, phosphorylation site prediction, and other techniques, we identify potential kinase targets. This is applied in tumor research, pathway analysis, and drug development.
4. Phosphorylation Dynamics Studies
Using time-resolved mass spectrometry (Time-resolved Phosphoproteomics), we monitor dynamic changes in phosphorylation during signal transduction. This is suitable for studies on drug stimulation, cell cycle regulation, and more.
Why Choose MtoZ Biolabs
With extensive experience in post-translational modification (PTM) proteomics research, MtoZ Biolabs provides high-quality phosphoproteomics solutions for researchers.
✅ Ultra-High Sensitivity and Specificity: Utilizing the latest HRMS technology, we can precisely identify low-abundance phosphorylated peptides.
✅ Optimized Enrichment Strategies: We employ various phosphorylation enrichment techniques, including IMAC and TiO₂, to enhance the detection efficiency of modified peptides.
✅ Multidimensional Quantitative Analysis: We support quantification strategies such as TMT and Label-free, ensuring flexibility for different experimental needs.
✅ Professional Bioinformatics Support: Using the latest databases and algorithms, we provide accurate data interpretation.
✅ Custom Experimental Solutions: We offer personalized experimental design and analysis tailored to your specific research requirements.
✅ Efficient Project Delivery: From sample receipt to data analysis, we ensure strict quality control throughout the entire process to guarantee the reliability of the data.
Applications
Phosphoproteomics has wide applications in both basic research and clinical translation, including:
✅ Cancer Research: Using phosphoproteomics solutions to identify cancer-related phosphorylation signaling pathways and discover potential biomarkers.
✅ Neuroscience: Analyzing neuronal phosphorylation regulatory mechanisms to study neurodegenerative diseases such as Alzheimer's.
✅ Metabolic Disease Research: Revealing signaling pathways associated with metabolic disorders like diabetes.
✅ Drug Development: Screening kinase inhibitor targets and optimizing drug screening strategies.
✅ Autoimmune Diseases: Investigating abnormal signaling pathways in autoimmune diseases to provide a foundation for precision medicine.
FAQ
Q1. Why must samples be processed quickly in phosphoproteomics studies?
A: Phosphorylation is a dynamic and reversible modification process. Even after cells are harvested, active phosphatases (e.g., PP1/PP2A) and kinases remain active. Studies have shown that after 30 minutes at room temperature, liver tissue samples show a 60% decrease in p-AKT (S473) levels.
General Solutions:
Q2. Why do phosphopeptides require special enrichment?
A: Technical Challenges:
Mainstream Enrichment Methods:
| Method | Principle | Applicable Scenarios |
|---|---|---|
| Metal Affinity Chromatography | Phosphate groups bind with TiO₂ | Broad-spectrum enrichment for a wide range of phosphorylated peptides |
| Antibody Immunoprecipitation | Targets specific phosphorylation sites (e.g., p-Tyr) | Targeted research focusing on specific modifications or sites |
| Chemical Modification | β-elimination reaction labels phosphate groups | Quantitative analysis of phosphorylation levels |
Phosphoproteomics is a key tool for uncovering the dynamic regulation of life processes. MtoZ Biolabs is dedicated to providing cutting-edge phosphoproteomics solutions to researchers worldwide, supporting pioneering scientific exploration. If you require precise and comprehensive phosphoproteomics analysis, feel free to contact us and begin a new chapter in your research!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Post-Translational Modifications Proteomics Service
How to order?







