Phosphoproteomics Service
Phosphoproteomics is a discipline dedicated to studying protein phosphorylation modifications and their roles in cellular signal transduction, metabolic regulation, and disease pathogenesis. Phosphorylation is a crucial post-translational modification (PTM) that primarily occurs on serine (Ser), threonine (Thr), and tyrosine (Tyr) residues, catalyzed by protein kinases, affecting protein activity, stability, interactions, and localization. The core principle of phosphoproteomics relies on high-resolution liquid chromatography-mass spectrometry (LC-MS/MS), combined with phosphopeptide enrichment strategies such as titanium dioxide (TiO₂), immobilized metal affinity chromatography (IMAC), and metal oxide affinity chromatography (MOAC) to precisely identify and quantify phosphorylation sites, thereby revealing dynamic intracellular signaling networks. Phosphoproteomics service provides high-resolution insights into protein modifications, facilitating the discovery of biomarkers and drug targets, accelerating disease diagnosis and therapeutic strategy development, and enhancing research efficiency and data reliability through its high throughput and sensitivity.
Higgins, L. et al. Biochemical Journal, 2023.
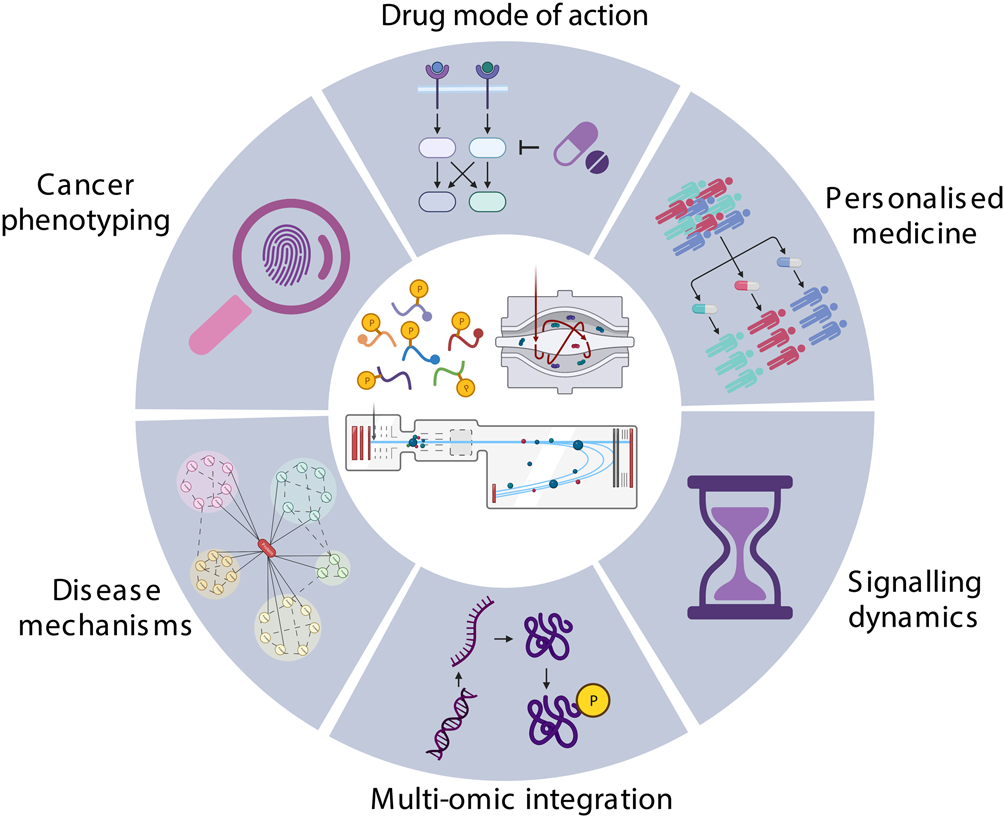
Figure 1. Application of Phosphoproteomics in Cancer Research.
Services at MtoZ Biolabs
Leveraging high-resolution mass spectrometry technology, MtoZ Biolabs provides phosphoproteomics service for high-throughput analysis of phosphorylation sites and dynamic modifications in complex biological samples. Our comprehensive workflow includes sample preparation, protein extraction, enzymatic digestion, phosphopeptide enrichment, LC-MS/MS detection, and bioinformatics analysis, ensuring precise phosphorylation site identification and high-quality quantitative data. By comprehensively analyzing phosphorylation modification patterns, we reveal dynamic changes in signaling pathways, key kinase activity regulation, and functional impacts of post-translational modifications, supporting disease research, drug target screening, and cellular signaling pathway analysis.
Service Advantages
1. State-of-the-Art Mass Spectrometry Technology
MtoZ Biolabs' phosphoproteomics service employs the latest high-resolution mass spectrometers (such as LC-MS/MS), significantly enhancing the detection sensitivity of phosphorylation sites. This enables precise analysis in scenarios with limited sample amounts or low-abundance model systems.
2. Efficient Phosphorylation Site Enrichment and Quantitation
Targeting key phosphorylation sites, MtoZ Biolabs utilizes high-affinity enrichment reagents and quantitative mass spectrometry methods to accurately identify and quantify low-abundance phosphorylation sites. This approach facilitates the elucidation of complex signaling networks.
3. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
4. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all phosphoproteomics data, providing clients with a comprehensive data report.
5. Customized Solutions
Tailored experimental design based on research objectives, including specific signaling pathway analysis, key kinase identification, and phosphorylation dynamics studies, supporting precision medicine, cancer research, and new drug development.
Applications
1. Disease Research
In cancer, autoimmune diseases, and neurodegenerative disorders, analyzing protein phosphorylation patterns reveals abnormalities in disease-related signal transduction and molecular mechanisms, helping to identify potential biomarkers and therapeutic targets.
2. Drug Development
Phosphoproteomics service can be used to assess the impact of drugs on cellular signaling pathways, optimize drug design, enhance drug efficacy, and reduce side effects.
3. Integration of Basic Research and Clinical Applications
This analysis can provide high-resolution information on protein modifications, facilitating the translation of basic research findings into clinical applications.
4. Cell Signaling Pathway Research
Phosphorylation is a key regulatory mechanism in signal transduction. Phosphoproteomics service enables the analysis of critical biological processes, including growth factor signaling, immune response, metabolic regulation, and cell cycle control, revealing the role of protein phosphorylation in cellular biology.
5. Precision Medicine and Personalized Treatment
By profiling individual patients' phosphoprotein expression patterns and integrating multi-omics data, phosphoproteomics can facilitate disease subtyping, optimize cancer-targeted therapies and immunotherapies, and enhance personalized medical strategies for improved treatment outcomes.
Case Study
1. Quantitative Phosphoproteomic Profiling of Mouse Sperm Maturation in Epididymis Revealed Kinases Important for Sperm Motility
This study aimed to investigate the maturation process of mouse sperm in the epididymis using quantitative phosphoproteomics analysis, identifying key kinases associated with sperm motility. The research subjects included sperm samples collected from the caput, corpus, and cauda regions of the mouse epididymis. High-resolution mass spectrometry combined with phosphopeptide enrichment strategies was employed to quantitatively analyze phosphoproteins at different maturation stages, followed by bioinformatics-based functional analysis. The results revealed significant changes in phosphorylation levels of multiple proteins during sperm maturation, particularly those related to flagellar movement, energy metabolism, and signal transduction. Further analysis indicated that sperm maturation is accompanied by dynamic regulation of key kinases, with kinases such as PKA, PKC, and CAMK exhibiting enhanced phosphorylation specifically in caudal sperm, suggesting their critical role in regulating sperm motility. Additionally, several potential phosphorylation sites associated with sperm motility were identified, and functional validation experiments confirmed their regulatory function in sperm flagellar dynamics. The study concluded that phosphorylation plays a crucial role in sperm maturation, with specific kinase regulation potentially being a key mechanism for sperm acquiring motility competence, providing novel molecular insights into male reproductive physiology and related disorders.
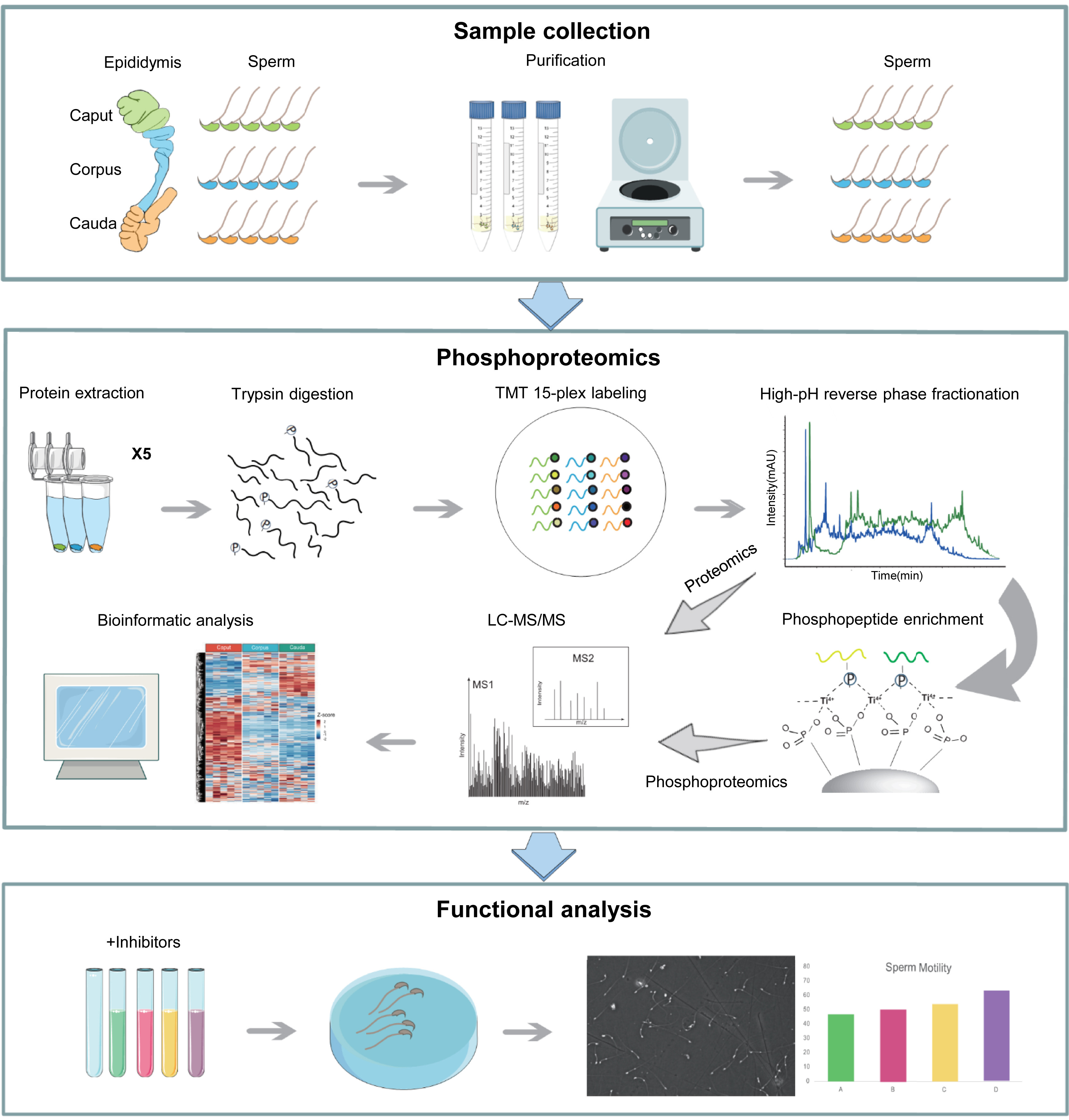
Zhang, X Z. et al. Molecular & Cellular Proteomics, 2024.
Figure 1. Workflow of Phosphoproteomic Analysis of Maturing Mouse Sperm from Different Epididymal Regions.
FAQ
Q: How does the instability of the phosphate group during collision-induced dissociation affect the accurate identification and localization of phosphorylation sites?
A: Specific mass spectrometry techniques such as higher-energy collision-induced dissociation (HCD) or electron-transfer higher-energy collision-induced dissociation (EThcD) can be employed to enhance the retention and recognition efficiency of phosphorylation sites. Additionally, combining multiple fragmentation methods and utilizing specialized software algorithms can more accurately resolve phosphorylation sites, reducing identification errors caused by the loss of phosphate groups, thereby ensuring the accurate localization of phosphorylation sites.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Relevant Liquid Chromatography and Mass Spectrometry Parameters
4. The Detailed Information of Phosphoproteomics
5. Mass Spectrometry Image
6. Raw Data
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Post-Translational Modifications Proteomics Service
How to order?