Olink Proteomics Analysis Service
Olink proteomics analysis based on proximity extension assay (PEA) technology enables precise quantification and relative comparison of up to 3,000 proteins in a single analysis. The core of PEA technology is to convert protein expression information into amplifiable DNA signals through spatial neighbor binding of antibody pairs and specific extension of DNA sequences, thereby achieving efficient detection of proteins. By utilizing specific antibody pairs and DNA tags, combined with highly sensitive qPCR or NGS detection, olink proteomics analysis service can provide high-resolution protein expression data and relative quantification results. MtoZ Biolabs offers olink proteomics analysis service with high specificity and sensitivity for high-throughput, simultaneous protein analysis.
Analysis Workflow
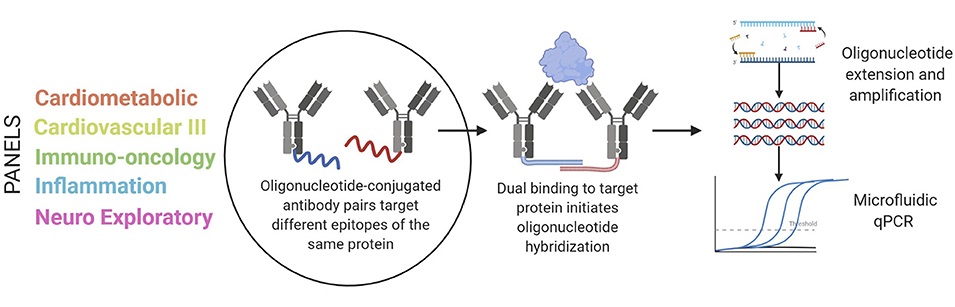
Carlyle B C. et al. Front Neurol. 2022.
1. Sample Pretreatment
Perform standardized preprocessing based on sample type to ensure protein integrity and stability.
2. Olink Panel Selection
Choose pre-designed thematic panels (e.g., oncology, cardiovascular, inflammation, neurology) or customized panels based on research needs.
3. Antibody Reaction and DNA Tag Binding
React samples with antibody reagents, antibodies specifically recognize target proteins and complete the proximity extension reaction through DNA tags.
4. Amplification and Data Generation
Detect DNA tags via qPCR or NGS, generating quantitative signals proportional to protein concentrations.
5. Data Analysis and Interpretation
Use specialized software to standardize and statistically analyze the generated data, providing protein expression levels and differential comparison results.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced olink proteomics analysis platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all olink proteomics analysis data, providing clients with a comprehensive data report.
Applications
Disease Molecular Subtyping
Reveal disease-associated protein networks to support diagnosis and typing research for precision medicine.
Novel Target Discovery
Identify key regulatory proteins involved in disease processes, providing a scientific basis for new drug development.
Biomarker Validation
Validate the expression characteristics of candidate biomarkers in different samples, supporting early diagnosis and treatment monitoring.
Complex Biological Systems Research
Investigate the roles of metabolism, immunity, and signaling pathways in complex biological problems.
Drug Development and Evaluation
Quantitatively assess the impact of drugs on target proteins, providing data support for efficacy and toxicity studies.
FAQ
Q. What Are the Advantages and Limitations of Olink Proteomics Analysis Compared to Other Proteomics Methods (e.g., LC-MS/MS, ELISA)?
Advantages:
1. High Sensitivity
Olink uses PEA technology, achieving detection sensitivity at the pg/mL level, making it ideal for analyzing low-abundance proteins.
2. High Specificity
Dual antibody recognition and complementary extension of DNA tags significantly reduces nonspecific signals, ensuring highly reliable results.
3. High Throughput
Detects up to 3,000 proteins in a single experiment, far exceeding traditional ELISA, and the data consistency is better than LC-MS/MS.
4. Low Sample Volume Requirement
Requires only 10 µL of sample for comprehensive proteomic analysis, making it particularly suitable for precious or rare samples.
5. Rapid Data Generation
Combined with qPCR or NGS technology, data processing is fast and results are formatted for easy subsequent analysis.
Limitations:
1. Limited Protein Detection Range
Olink relies on predefined antibody panels, restricting the analysis to a fixed set of proteins. It cannot be freely expanded to target proteins outside the panel.
2. Lack of Post-Translational Modification (PTMs) Information
Olink focuses solely on protein expression levels and cannot directly detect modifications such as phosphorylation or acetylation, requiring complementary methods like LC-MS/MS.
3. High Cost
Olink is relatively expensive, especially in large-scale studies, which may impose budget constraints.
Olink proteomics analysis service is highly suited for high-throughput, low-abundance protein detection, and biomarker discovery, particularly in precision medicine research. However, for unknown protein exploration or research requiring PTMs analysis, it is recommended to combine Olink with other proteomics approaches like LC-MS/MS.
Case Study
This study utilized olink proteomics analysis to conduct a longitudinal investigation of dynamic changes in lung protein expression in mild to moderate COVID-19 patients. The findings revealed that proteins associated with immune response, inflammation regulation, and tissue repair remained persistently expressed in the lungs for months post-infection. These results reveal a possible long-term lung repair process after clinical recovery from COVID-19 and provide potential biomarkers and intervention targets.
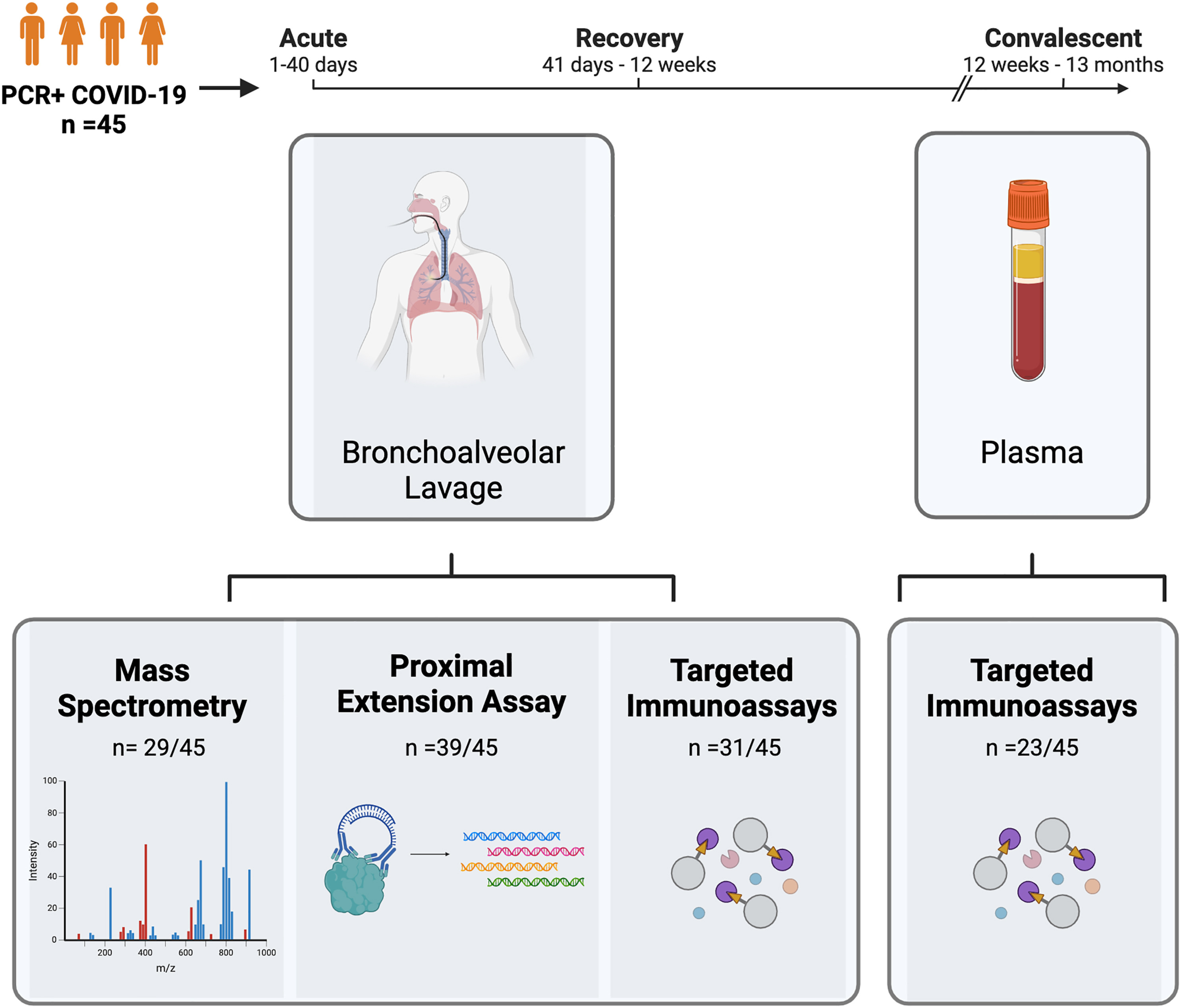
Kanth S M. et al. Cell Reports Medicine. 2024.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







