Next-Generation De Novo Sequencing Algorithms
-
Utilizing spectral similarity metrics (e.g., cosine similarity scores);
-
Integrating structural conservation and homologous sequence alignment information;
-
Supporting the simultaneous inference of peptide isomers to enhance the robustness of final identifications.
De Novo sequencing, defined as the de novo inference of peptide amino acid sequences, is a critical technique in proteomics for the identification of unknown proteins, post-translationally modified peptides, and antibody fragments. In contrast to database search-based approaches, De Novo sequencing operates independently of reference databases, rendering it particularly effective for complex scenarios such as non-model organisms, incomplete database coverage, post-translational modifications, and highly variable samples. Nevertheless, conventional De Novo algorithms (e.g., PEAKS, Novor, PepNovo) continue to face challenges in resolving ambiguities in fragment ion spectra, identifying long peptide sequences accurately, and detecting low-abundance signals. To address these limitations, next-generation De Novo sequencing algorithms have introduced systematic advancements in model architectures, spectral representations, and training paradigms. This article focuses on these technological developments, highlighting the key innovations and practical implementations of next-generation algorithms.
Brief Introduction to the Development of De Novo Sequencing
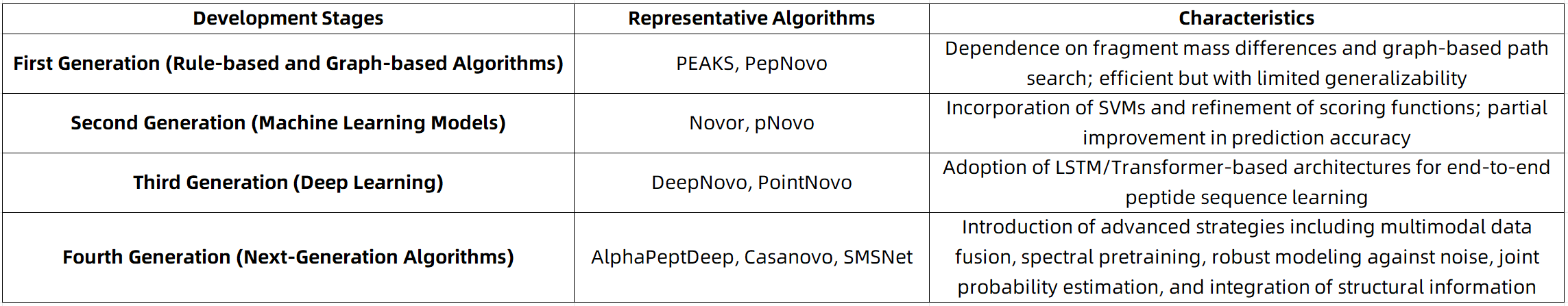
Key Technological Innovations in Next-Generation De Novo Sequencing Algorithms
1. Integration of Transformer Architecture
Compared to traditional recurrent neural networks (such as LSTM), Transformer architectures exhibit superior capabilities in modeling long-range dependencies. This enables them to concurrently capture both fragment distribution and contextual relationships across the entire mass spectrum. Representative models include:
(1) AlphaPeptDeep: developed jointly by Microsoft Research and the Max Planck Institute, this model integrates spectral and sequence-level information through joint modeling;
(2) Casanovo: proposed by the Facebook research team, utilizes a Transformer architecture to directly predict peptide sequences from raw MS/MS fragment ion vectors;
(3) pDeep3: combines predicted spectra with reverse scoring strategies to re-rank candidate peptide sequences.
2. Multi-Modal Data Input
While conventional De Novo algorithms rely solely on spectral data, next-generation models incorporate diverse input modalities, including:
(1) MS/MS spectral data (represented as vectors or graph-based structures);
(2) Sequence context features (e.g., amino acid residue frequencies, species-specific background information);
(3) Experimental metadata (such as fragmentation methods and instrument types);
(4) Optional priors on peptide structure and post-translational modifications.
These enhancements not only improve overall prediction accuracy but also significantly enhance the identification of non-standard modifications and heterogeneous peptide sequences.
3. Spectrum Pretraining and Transfer Learning
Inspired by developments in natural language processing (e.g., BERT and GPT), several models leverage large-scale pretraining on spectral data to facilitate transfer learning:
(1) Models are pre-trained on millions of unlabeled peptide-spectrum matches using unsupervised learning;
(2) High predictive accuracy is retained even when applied to tasks with limited labeled data;
(3) The framework can be adapted across diverse mass spectrometry platforms and enzymatic digestion strategies.
4. Re-Ranking of Candidate Peptide Sequences
Post-prediction re-ranking modules have been introduced to refine the confidence scoring of multiple predicted sequences:
Application Advantages: Enhanced Intelligence, Sensitivity, and Suitability for Highly Complex Biological Samples
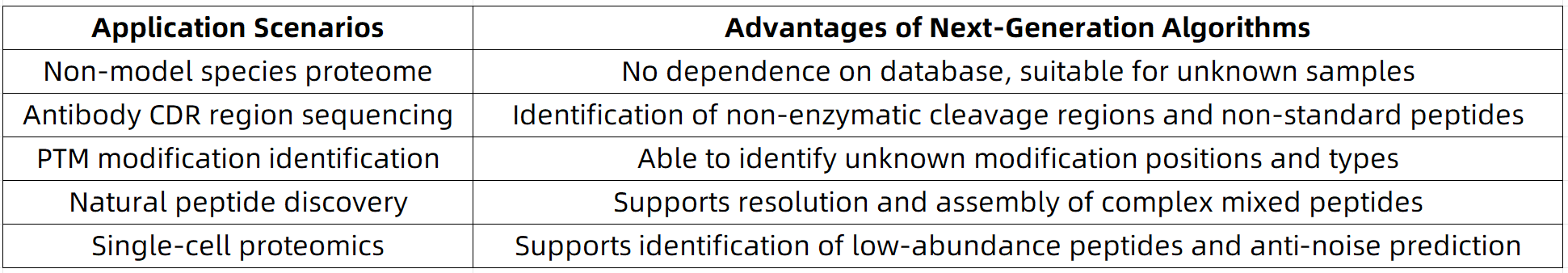
MtoZ Biolabs’ Intelligent Peptide Analysis Platform
To accommodate a broad range of research objectives, MtoZ Biolabs has developed an intelligent De Novo sequencing platform that integrates multiple algorithmic strategies, offering the following capabilities:
1. System Architecture
(1) Algorithm Engine: Incorporates DeepNovo, AlphaPeptDeep, Casanovo, and proprietary modules;
(2) Data Processing: Supports DIA/DDA spectral interpretation, multi-protease digestion, and post-translational modification identification;
(3) Sequence Validation: Enables confirmation via synthetic peptide spectral matching;
(4) Functional Annotation: Integrates homology-based searches and structural modeling to facilitate biological interpretation.
2. Service Use Cases
(1) Antibody sequencing (monoclonal, humanized antibodies, and scFv fragments)
(2) Proteomic analysis of ultra-low abundance samples (e.g., exosomes, cerebrospinal fluid)
(3) Discovery and activity prediction of novel endogenous peptides
(4) Proteome profiling of non-model organisms
With the increasing integration of artificial intelligence into proteomic research, De Novo sequencing has transitioned from a “proof-of-concept” approach to a “frontline analytical strategy.” Advances in next-generation algorithms are significantly expanding our capacity to characterize unknown peptides, protein variants, and modification patterns. At MtoZ Biolabs, we are committed to accelerating the convergence of AI and proteomics, delivering high-throughput, high-fidelity, and experimentally validated De Novo sequencing solutions. We welcome research collaborations and inquiries aimed at advancing next-generation protein characterization.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







