Neddylation Proteomics Service
Neddylation proteomics analysis is a high-resolution proteomics technique dedicated to the identification and characterization of protein neddylation modifications. Neddylation is a ubiquitin-like post-translational modification in which NEDD8 (Neural precursor cell expressed developmentally downregulated protein 8) is covalently attached to lysine residues of substrate proteins through an E1-E2-E3 enzymatic cascade. This modification regulates protein stability, activity, and interaction with ubiquitin ligase complexes. It plays a particularly critical role in modulating the activity of Cullin-RING E3 ligases (CRLs), serving as a key regulatory node in the cellular protein degradation pathway.
Neddylation proteomics service is widely applied in studies of the ubiquitin-proteasome system, signal transduction network analysis, tumorigenesis mechanism exploration, and regulation of autophagy and inflammatory responses. It also supports the identification of potential drug targets related to neddylation and contributes to the development of targeted anti-cancer and anti-inflammatory therapeutic strategies.
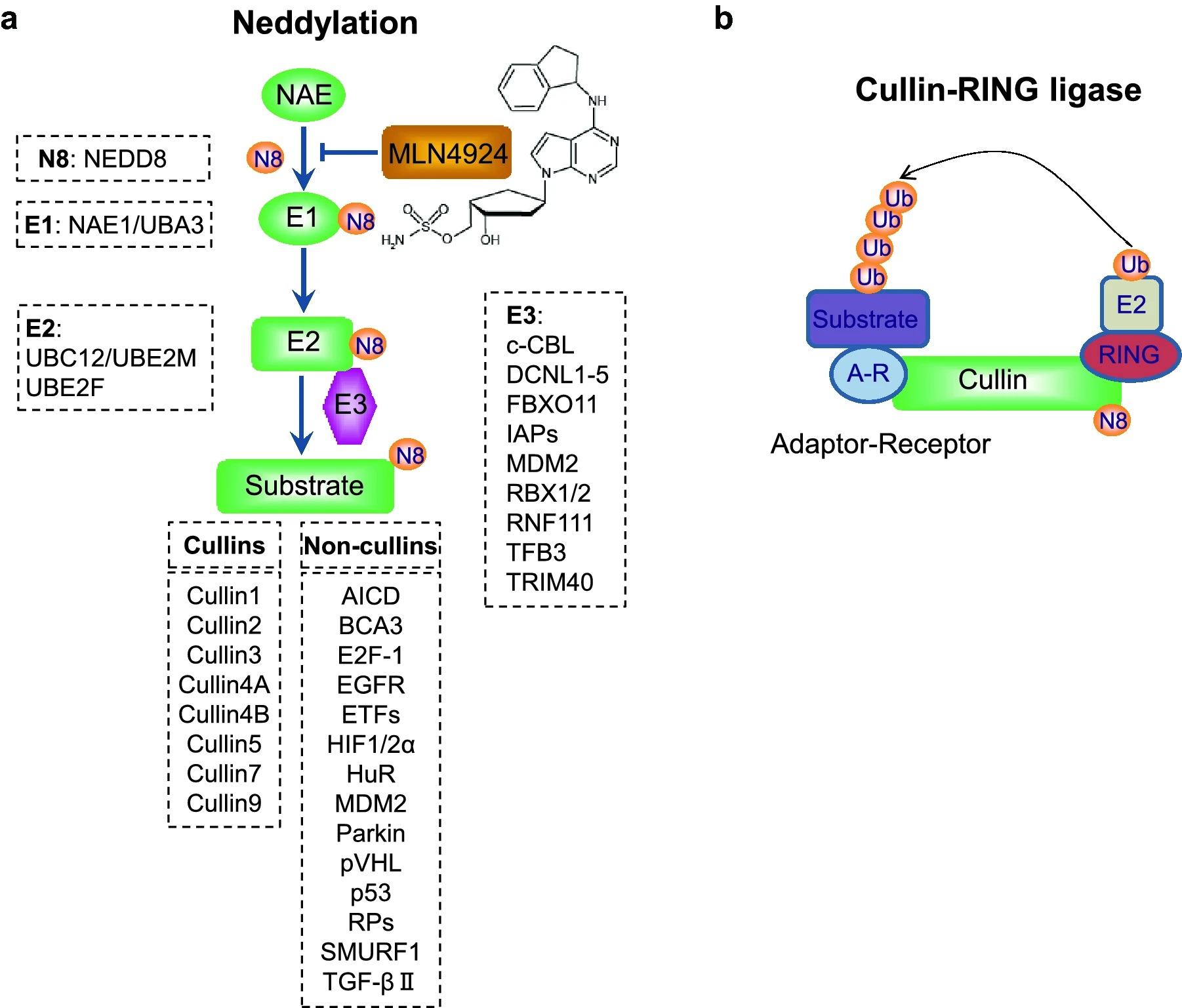
Zhou, Q Y. et al. Cell & Bioscience, 2021.
Figure 1. The Process of Neddylation Modification.
Services at MtoZ Biolabs
Based on advanced analytical platforms and customized workflows for sample preparation and NEDD8-modified peptide enrichment, the neddylation proteomics service offered by MtoZ Biolabs enables highly sensitive analysis of protein neddylation modifications. This service supports systematic profiling of neddylated proteins in complex samples such as cells and tissues, accurately identifying modification sites and corresponding peptide sequences. It provides high-quality data including peptide sequences, NEDD8 chain lengths, and quantitative changes, facilitating comprehensive analysis and functional interpretation of post-translational modifications. Commonly used methods in this service include the following:
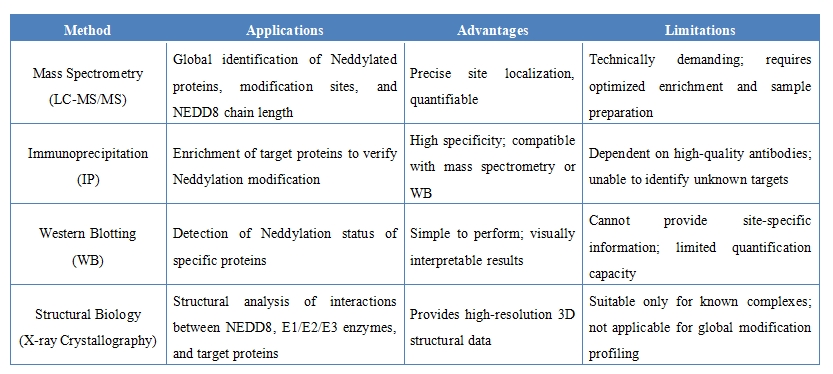
Analysis Workflow
1. Protein Extraction and Digestion
Total proteins are extracted from cell or tissue samples, followed by reduction, alkylation, and enzymatic digestion. This preserves neddylation modification information and prepares the sample for downstream analysis.
2. Enrichment of Modified Peptides
NEDD8-modified peptides are selectively enriched using specific antibody-based or chemical labeling strategies, significantly improving the detection efficiency of low-abundance modifications.
3. Multi-Technology Detection
Depending on research needs, mass spectrometry, Western blotting, or structural biology approaches can be employed to precisely identify modification sites, peptide sequences, and NEDD8 chain lengths, and to validate the presence, distribution, and functional impact of the modification.
4. Data Analysis and Result Reporting
Through database searching and algorithmic processing, results are generated including neddylation sites, quantitative changes, modification spectra, and functional annotations to support downstream mechanistic studies.
Sample Submission Suggestions
1. Sample Types
Various sample types are supported, including cells, tissues, body fluids, and purified proteins. It is recommended to select cell lines or treatment conditions with high neddylation activity to improve modification detection efficiency.
2. Buffer System
Avoid using buffers containing high salt, SDS, glycerol, or strong reducing agents that may affect modification stability or compatibility with mass spectrometry. Buffer optimization guidance is available upon request.
3. Sample Transport
Samples should be frozen at –80°C and shipped on dry ice to preserve the integrity of neddylation modifications and overall protein stability.
Service Advantages
1. High-Resolution Platform Support
Powered by advanced mass spectrometry systems such as Orbitrap, the service ensures highly sensitive and accurate identification and quantification of neddylation modifications.
2. Specific Enrichment Strategy
Combining anti-NEDD8 antibodies and chemical labeling approaches, the service effectively captures low-abundance modified peptides, improving both detection efficiency and specificity.
3. Flexible Detection Strategy Support
Depending on research objectives, a range of methods—such as quantitative mass spectrometry, immunological validation, and structural analysis—can be applied to accommodate both high-throughput screening and in-depth mechanistic studies.
4. One-Stop Service
Offering end-to-end support from sample processing to data interpretation, the service delivers comprehensive results including modification sites, peptide sequences, modification intensity, and functional annotation to support target discovery and mechanistic exploration.
Applications
1. Protein Degradation Pathway Research
Neddylation proteomics service can be used to elucidate how NEDD8 regulates the activity of Cullin-RING E3 ligases (CRLs), highlighting its essential role in the ubiquitin–proteasome system.
2. Signal Transduction Network Analysis
Through quantitative analysis, this service helps identify signaling nodes regulated by neddylation, uncovering its roles in pathways such as cell cycle control and stress responses.
3. Cancer and Inflammation Mechanism Research
The neddylation proteomics service supports the investigation of Neddylation’s regulatory functions in cell proliferation, DNA repair, and inflammatory processes, facilitating target discovery for cancer and immune-related diseases.
4. Post-Translational Modification Crosstalk
By integrating neddylation analysis with other modifications like ubiquitination and phosphorylation, this service reveals coordinated regulation within modification networks and their combined effects on protein function.
Case Study
1. Neddylation and MASLD: From Pathophysiology to Therapy
This study aims to systematically investigate the pathophysiological role of neddylation in metabolic-associated steatotic liver disease (MASLD) and its potential as a therapeutic target. The research involves liver tissues from MASLD patients, mouse MASLD models, and hepatic cell lines. Using proteomic analysis, expression profiling of key NEDD8 pathway enzymes, and functional intervention experiments, the researchers evaluated the impact of neddylation on disease progression. The results show that the neddylation pathway is abnormally activated during MASLD development, contributing to disrupted lipid metabolism, enhanced inflammatory responses, and hepatocellular stress—primarily through regulation of key metabolic pathways and transcription factor stability. Treatment with the neddylation inhibitor MLN4924 significantly alleviated hepatic lipid accumulation, reduced inflammation, and reversed liver tissue damage. The study concludes that neddylation plays a critical regulatory role in the pathogenesis of MASLD and that targeting this modification pathway may offer a promising therapeutic strategy.
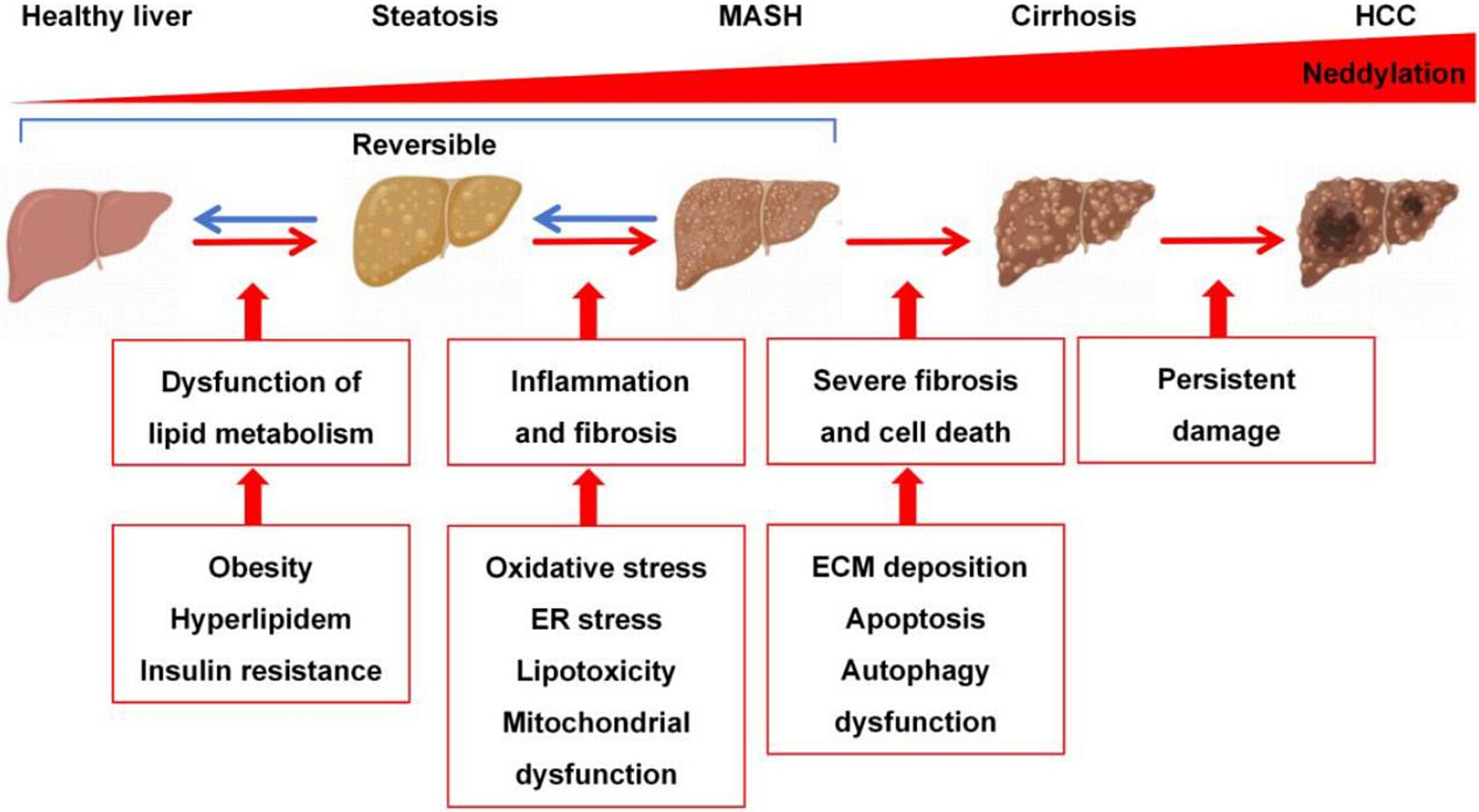
Lin, W J. et al. Liver International, 2025.
Figure 2. Neddylation in the Progression of MASLD.
2. Global Site-Specific Neddylation Profiling Reveals that NEDDylated Cofilin Regulates Actin Ddynamics
This study aimed to elucidate the role of neddylation in regulating actin dynamics through global site-specific neddylation proteomics, with a particular focus on the functional modulation of the cofilin protein. Endogenous proteins from human cells were analyzed using a combination of NEDD8-modified peptide enrichment strategies and high-resolution mass spectrometry to generate a comprehensive neddylation modification map. Cofilin was identified as a key neddylation target. The results revealed that site-specific neddylation of cofilin inhibits its binding to F-actin, thereby affecting actin filament depolymerization and cell migration. Mutation experiments further confirmed that this modification regulates the functional state of cofilin and contributes to cytoskeletal stability. The study concludes that neddylation regulates actin dynamics by modulating cofilin activity, serving as a crucial mechanism for maintaining cell motility and morphology.
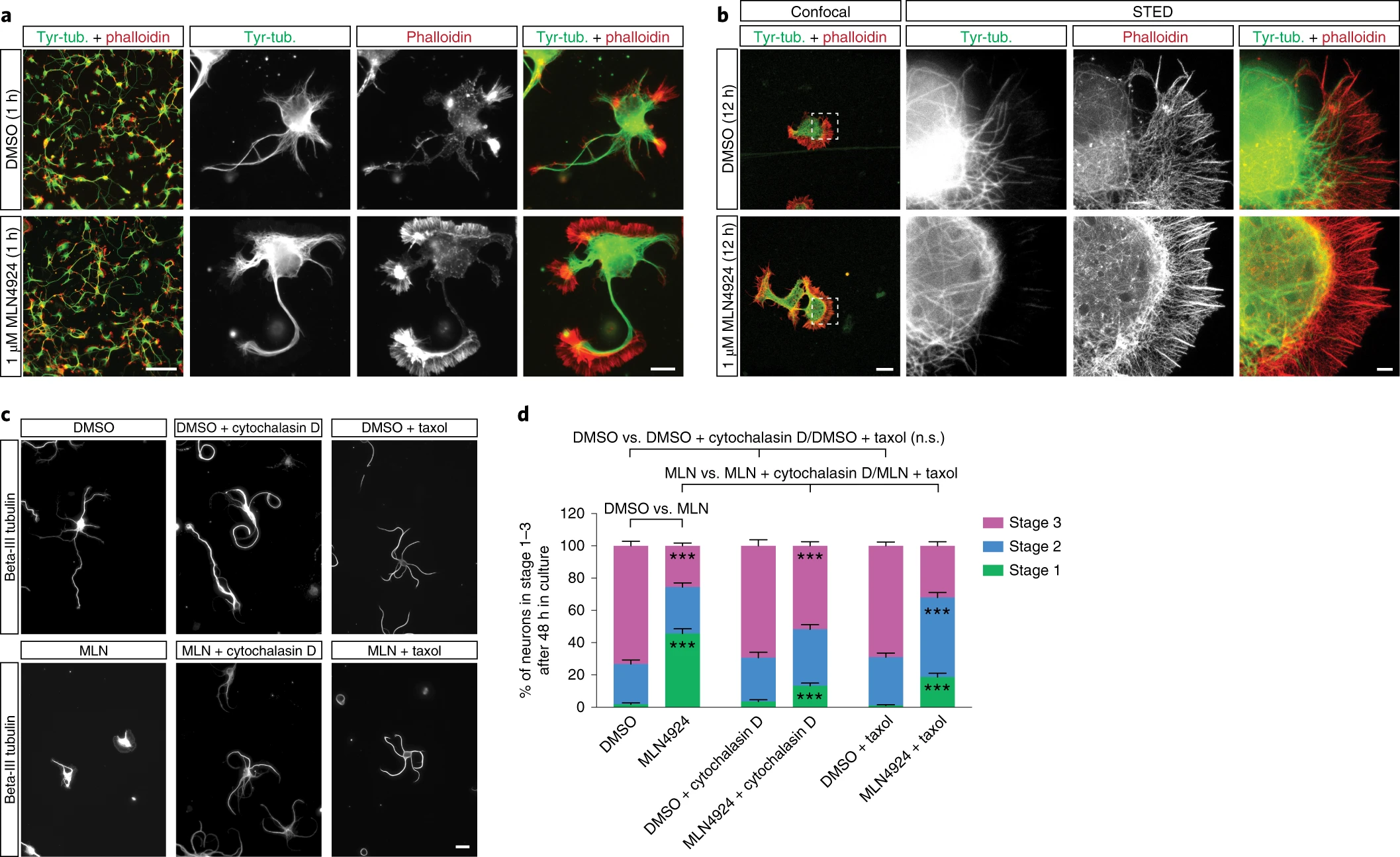
Vogl, A M. et al. Nature Structural & Molecular Biology, 2020.
Figure 3. Neddylation Modulates the Actin Cytoskeleton and Controls Neurite Outgrowth in Neurons.
FAQ
Q1: Can Low-Abundance Neddylated Proteins Be Detected?
A1: Yes. By applying an optimized enrichment strategy and leveraging the high sensitivity of the Orbitrap mass spectrometry platform, we can effectively detect low-abundance neddylated proteins even in complex biological backgrounds.
Q2: Is Quantitative Analysis Available?
A2: Yes. Relative quantification of neddylation modifications is supported through strategies such as label-free quantification or TMT labeling, enabling comparison of modification levels across different treatment groups.
How to order?







