Multi-protein Complexes Structure Characterization Service
Multi-protein Complexes Structure Characterization Service refers to the use of cryo-electron microscopy (Cryo-EM) technology to perform high-resolution three-dimensional structural analysis, atomic model construction, and interface functional annotation of biological macromolecular complexes composed of multiple subunits. This service covers the entire workflow from sample preparation to structural modeling and is suitable for research innovation, mechanism validation, target evaluation, and structure-based drug design.
Multi-protein complexes are higher-order structural units composed of two or more functionally related protein subunits assembled through non-covalent interactions or partial covalent bonds. They serve as the basic functional units responsible for executing the majority of life activities within the cell, participating in essential biological processes such as signal transduction, gene transcription, DNA repair, protein degradation, and energy metabolism. The spatial arrangement of subunits, interaction interfaces, and dynamic conformational changes within the complex directly determine its functional characteristics. Resolving their atomic-level structures is critical for understanding molecular mechanisms, identifying disease targets, and developing novel therapeutic strategies.
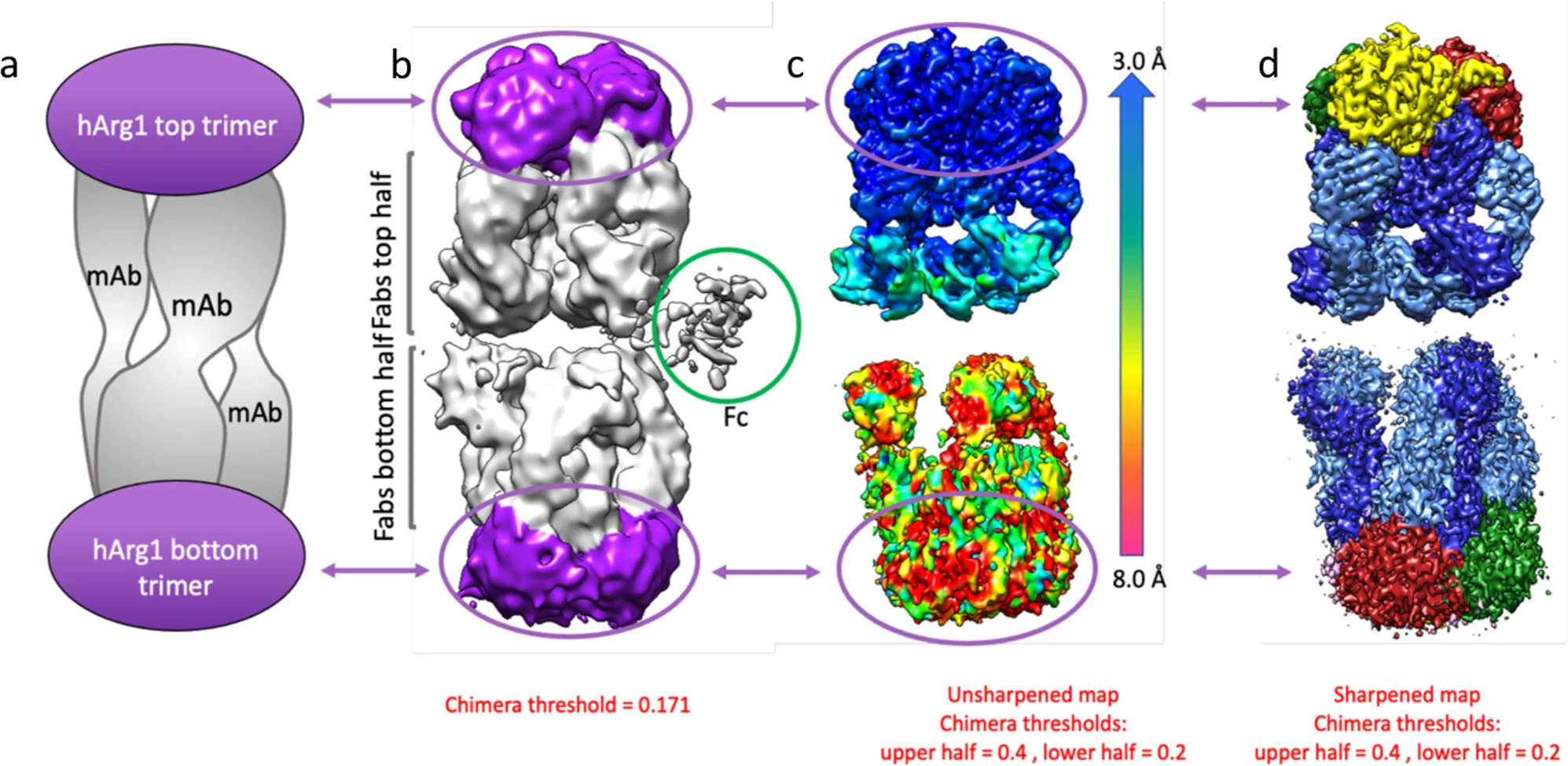
Palte RL. et al. Commun Biol. 2021.
Traditional X-ray crystallography and nuclear magnetic resonance (NMR) have significant limitations in studying the structures of multi-protein complexes, such as difficulties in sample preparation, crystallization barriers, and the inability to resolve dynamic regions. With the advancement of Cryo-EM technology, scientists are now able to resolve high-resolution three-dimensional structures of multi-protein complexes under near-physiological conditions without the need for crystallization.
Leveraging an advanced cryo-EM platform, MtoZ Biolabs provides the Multi-protein Complexes Structure Characterization Service assisting in the analysis of complex assembly mechanisms, subunit interaction interfaces, functional dynamic changes, and regulatory mechanisms. This service can offer precise structural support for basic research, mechanism exploration, structure-based drug design (SBDD), and molecular engineering optimization.
Analysis Workflow
The main workflow of the Multi-protein Complexes Structure Characterization Service is as follows:
1. Sample Preparation and Complex Assembly
Express and purify individual subunits, assemble the complex, and verify its composition and functional activity.
2. Cryo-Grid Preparation and Screening
Prepare high-quality cryo-grids, control ice thickness and particle dispersion. Assess particle distribution, integrity, and orientation uniformity.
3. High-Resolution Data Acquisition
Collect large amounts of high-quality image data under low-dose conditions using advanced cryo-EM platforms.
4. Image Processing and 3D Reconstruction
Perform automated particle picking, 2D/3D classification, multi-conformational recognition, and local refinement to generate high-resolution density maps.
5. Atomic Model Building and Structural Functional Annotation
Build complete atomic coordinate models based on the density maps, and annotate subunit interaction interfaces, functional binding sites, and dynamic transition pathways.
6. Result Delivery and Biological Interpretation
Provide standardized data packages, interface maps, and complete functional analysis reports to support scientific publication, patent application, and drug development.
Applications
The applications of the Multi-protein Complexes Structure Characterization Service include:
Signal Transduction Complex Analysis
Such as GPCR-G protein complexes, receptor-ligand-cofactor assemblies.
Transcription Regulation and Chromatin Remodeling Complexes
Such as mediator complex, polycomb complex, nucleosome remodeling complexes.
Protein Degradation System Structure Analysis
Such as E3 ubiquitin ligase-substrate complexes, proteasome-adaptor complexes.
DNA Repair and Replication Complex Structure Analysis
Such as MRN complex, nuclease-repair factor complexes.
Complex Studies for Vaccine and Biologic Development
Such as antigen-antibody-immune modulator complex structure design and optimization.
FAQ
Q. If a multi-protein complex exhibits heterogeneity or multiple conformations, can Cryo-EM distinguish and resolve them?
Yes. Through three-dimensional classification (3D classification) and local refinement techniques, Cryo-EM can effectively identify and separate different conformations or assembly states of particles, enabling high-resolution structure determination of the complex in various functional states—one of the key advantages for studying dynamic systems.
Q. Can Cryo-EM handle large complexes (>1 MDa) or highly flexible complexes?
Yes. Cryo-EM is particularly suited for studying large (>500 kDa), highly heterogeneous, or flexible complexes. For flexible regions, local masking and Focused Refinement strategies can be used to further improve local resolution and analysis quality.
Case Study
This study used Cryo-EM technology to analyze the key multi-protein complex structure in the initial assembly of mitochondrial iron-sulfur clusters ([2Fe-2S] clusters under anaerobic conditions, including a complex composed of cysteine desulfurase NFS1, auxiliary protein ISD11, adenylate sulfate reductase ACP, scaffold protein ISCU2 and regulatory protein frataxin (FXN). By capturing multiple reaction intermediate states, the study revealed how the sulfur atom was precisely transferred from Cys381 of NFS1 to the Cys138 site of ISCU2 through dynamic conformational changes, and that FXN played a key role in stabilizing the ISCU2 receptor site and promoting rapid sulfur transfer. This work systematically elucidates the molecular mechanism in de novo iron-sulfur cluster biosynthesis and provides important structural basis for understanding diseases related to iron-sulfur cluster assembly disorders, such as Friedreich's ataxia.
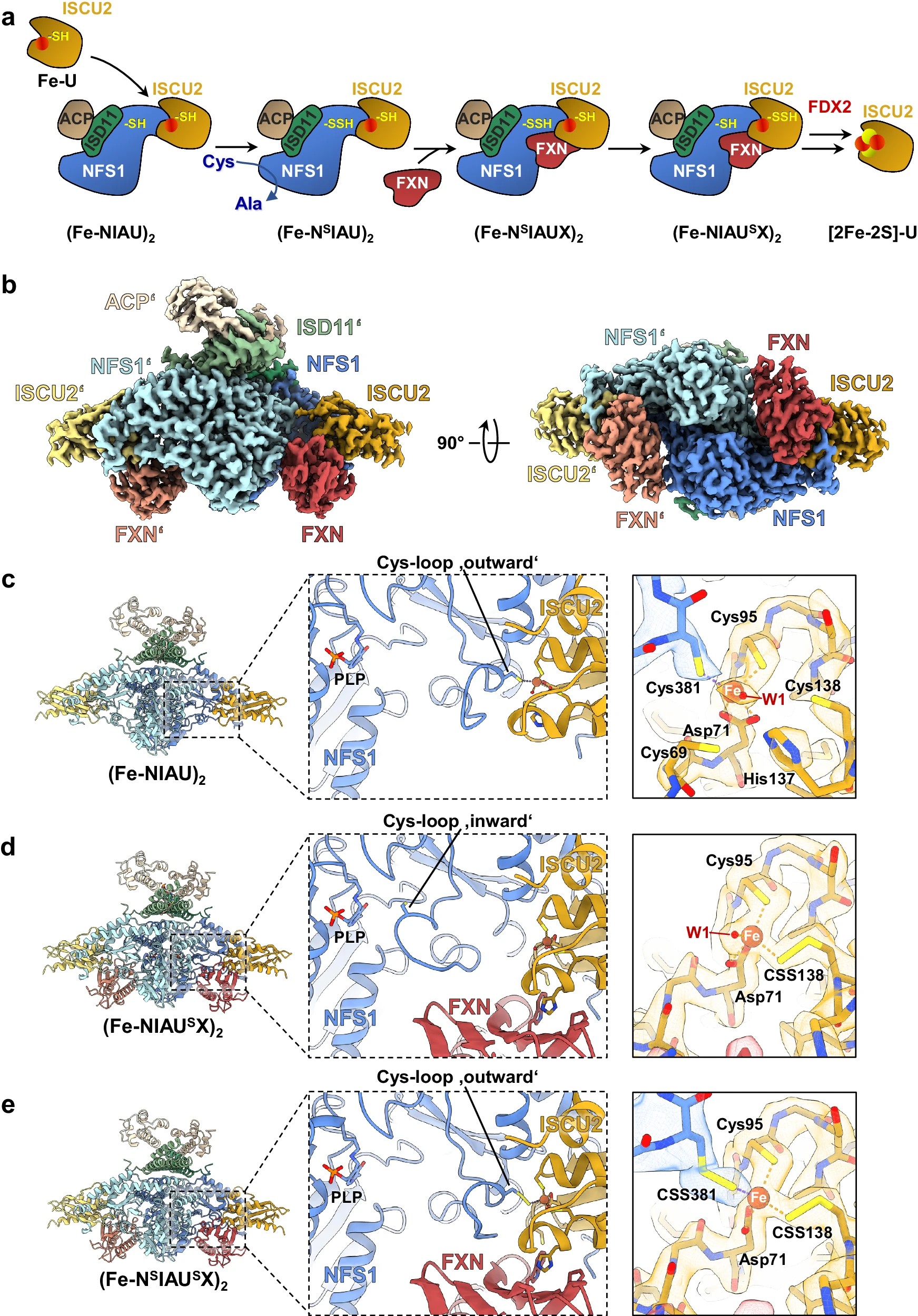
Schulz V. et al. Nat Commun. 2024.
How to order?







