Multi-omics Integration Analysis Service
- Biological significance: Focus on molecules with the most significant differential expression or node molecules in the core pathways.
- Statistical reliability: Select molecules with high significance based on p-values and FDR, and strong correlations.
- Functional annotation: Use databases like KEGG and GO to identify molecules known to be associated with the target disease or process.
- Network analysis: Use network centrality metrics (such as degree or betweenness), screen the molecules that have the greatest impact on system stability or function.
Multi-omics Integration Analysis Service integrates data from multiple omics layers, such as genomics, transcriptomics, proteomics, metabolomics, and more, to comprehensively analyze biological processes and uncover potential molecular mechanisms at a systems level. Compared to single-omics analysis, Multi-omics Integration Analysis provides a multidimensional view of the complex dynamic interactions between molecules, delivering comprehensive and in-depth biological insights. Leveraging advanced multi-omics analysis platforms, MtoZ Biolabs offers Multi-omics Integration Analysis Service which covers transcriptomics, proteomics, metabolomics, lipidomics and microbiomics, providing a full-process service from data acquisition to biological interpretation, empowering researchers to drive scientific innovation and industrial applications.
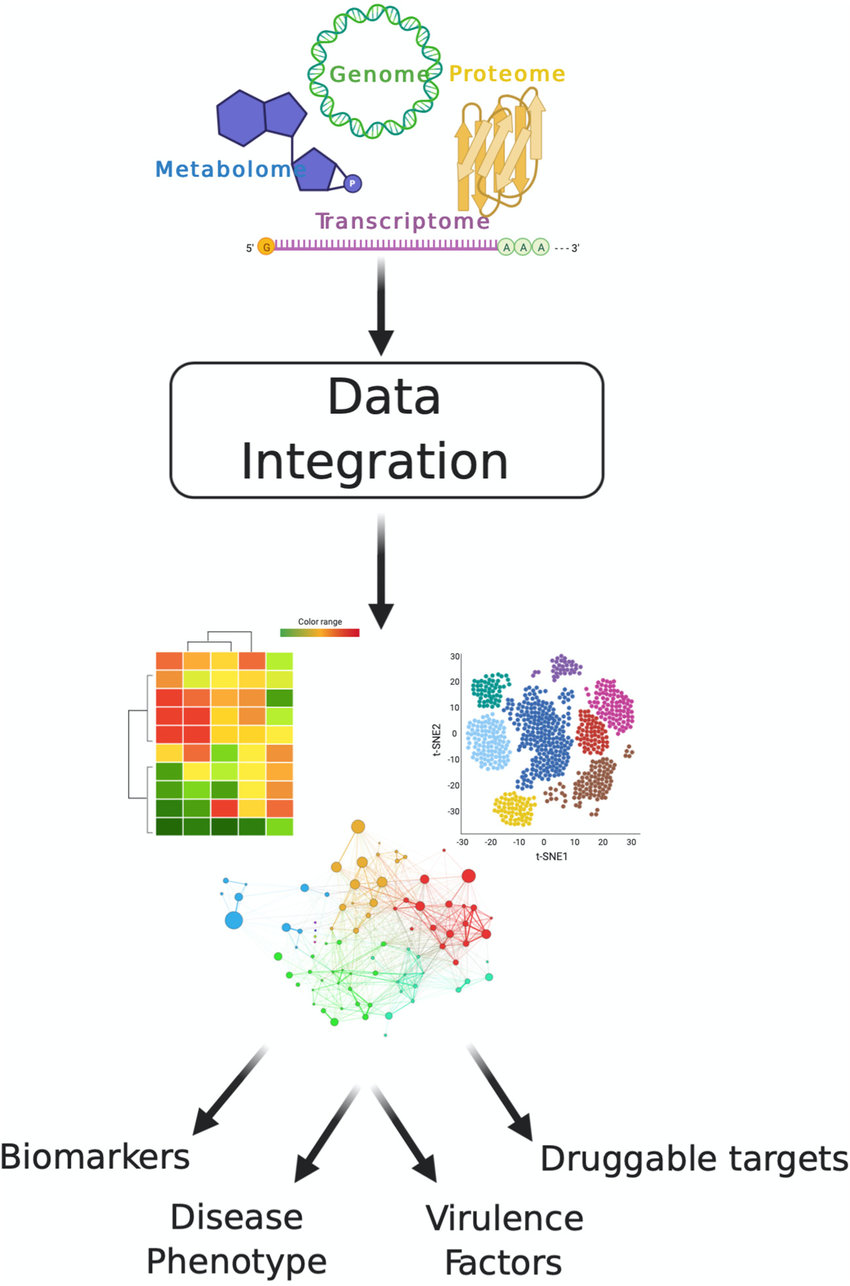
Ball B. et al. mBio. 2020.
Analysis Workflow
1. Experimental Design and Data Acquisition
Determine the appropriate omics layers based on research objectives (e.g., genomics + proteomics, transcriptomics + metabolomics). Acquire data using high-throughput technologies (e.g., next-generation sequencing) and mass spectrometry (e.g., LC-MS/MS).
2. Data Preprocessing and Quality Control
Conduct strict quality control on the collected data, remove background noise and outliers, and normalize the data.
3. Multi-omics Integration Analysis
Apply various statistical and computational approaches (e.g., multivariate statistics, network analysis, machine learning) to establish inter-omics relationships. Use correlation models (e.g., Pearson, Spearman) to uncover direct interactions between omics layers. Build multi-layer network models to reveal dynamic molecular interactions. Employ methods such as PCA, PLS-DA and random forest to extract features and identify key biomarkers. Conduct pathway enrichment analysis using functional databases (e.g., KEGG, GO, Reactome).
4. Result Interpretation and Report Generation
Output graphical results, such as network diagrams, heat maps, and PCA diagrams, and generate detailed analysis reports. Provide biological significance interpretation and follow-up experimental design suggestions.
Sample Submission Suggestions
Sample Types
We accept a variety of sample types, including but not limited to extracted RNA, microbial DNA, tissue lysates, serum, urine, etc.
Sample Quantity
Each omics analysis has different requirements. Specific needs can be customized based on the project. Please feel free to contact us for consultation before submitting your samples.
Applications
Disease Mechanism Research
Reveal multi-layered molecular mechanisms of complex diseases (e.g., cancer, metabolic disorders) to promote the development of precision medicine.
Biomarker Discovery
Screen biomarkers related to disease diagnosis, prognosis, or therapeutic response from multidimensional data through Multi-omics Integration Analysis.
Drug Development and Toxicity Evaluation
Analyze drug action targets, evaluate regulatory effects on molecular networks, predict potential toxic side effects, and support new drug development.
Systems Biology Research
Explore interactions among metabolic networks, signaling pathways, and gene regulatory networks.
Ecological and Microbial Studies
Investigate microbial community dynamics and their interactions with hosts or environments.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. With advanced equipment, a professional team, and strict quality control, MtoZ Biolabs is committed to providing researchers with high-quality Multi-omics Integration Analysis Services.
FAQ
Q. How to deal with the scale and dimensionality differences between different omics data? Is normalization or dimensionality reduction required?
Dealing with scale and dimensionality differences is a core challenge in multi-omics integration. First, normalization is necessary to eliminate the influence of technical noise and experimental conditions, such as using TPM/RPKM for transcriptome data and Z-score standardization for proteome and metabolome data. Second, according to the characteristics of the data, dimensionality reduction can simplify complex data and extract key information, such as using principal component analysis (PCA) or partial least squares discriminant analysis (PLS-DA) to extract the main patterns in high-dimensional data. At the same time, t-SNE or UMAP can be combined for visualization to facilitate understanding of the connection between omics.
Q. When multiple potential key molecules appear in the results of the integrated analysis, how to prioritize the targets of experimental verification?
Prioritize molecules that are closely related to the research objectives, have biological significance and statistical reliability. For instance:
Finally, prioritize candidate molecules in tiers and conduct functional validation experiments sequentially.
Related Services
This research utilizing Multi-omics Integration Analysis combined microbiome and metabolome data to investigate microbial and metabolic characteristics in patients with inflammatory bowel disease (IBD). The results demonstrated a significant reduction in gut microbial diversity in IBD patients, accompanied by notable alterations in inflammation-associated metabolites, such as short-chain fatty acids and bile acids. Multi-omics integration revealed correlations between specific microbes, such as Enterococcus faecium and Clostridium, and metabolites, highlighting their potential roles in the pathogenesis of IBD. This study provides potential biomarkers and targets for the diagnosis and treatment of IBD.
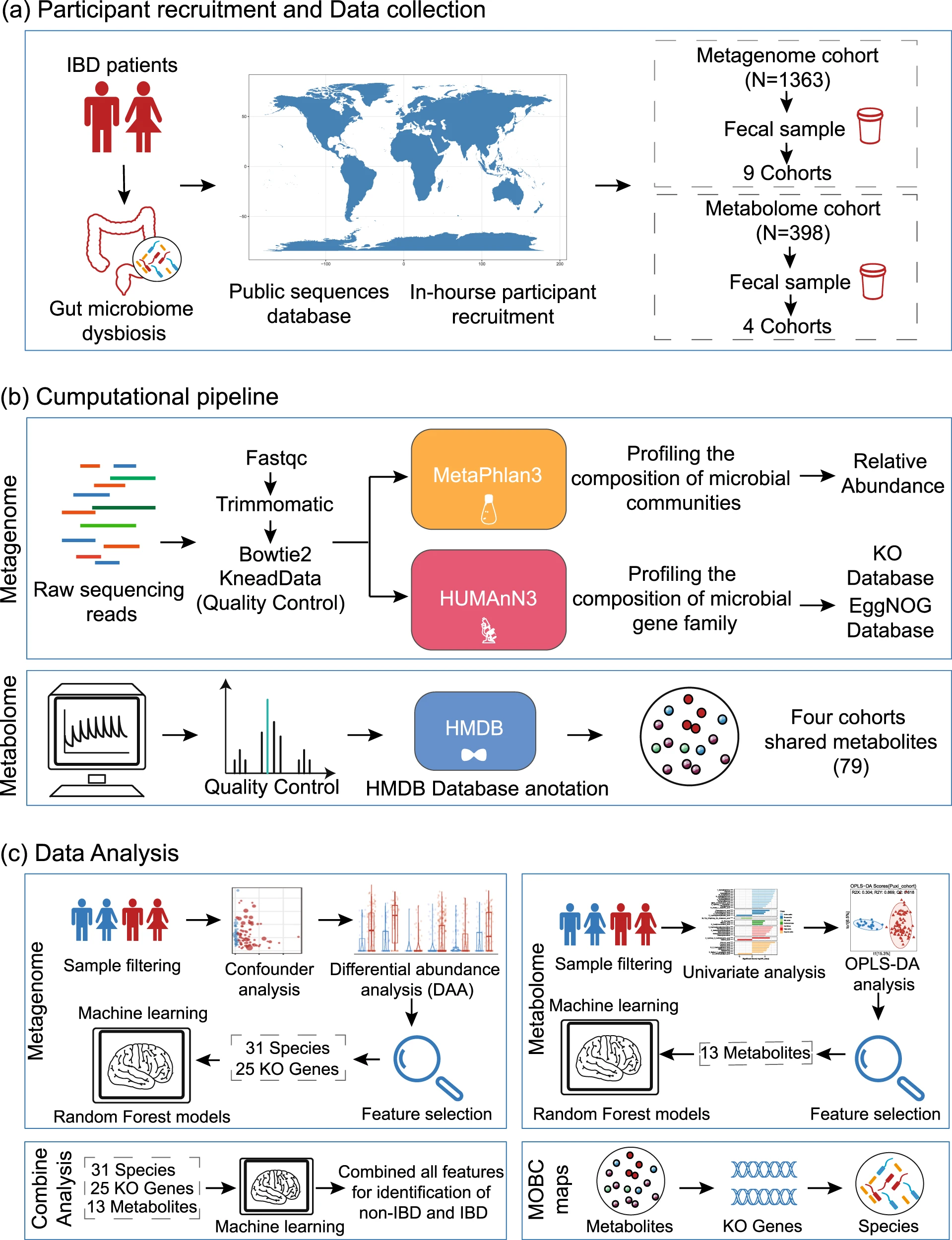
Ning L. et al. Nature Communications. 2023.
How to order?







