MS Based Proteomics Analysis Service
MS based proteomics analysis service uses mass spectrometry as the core technology to study the composition, structure and function of proteins in biological samples. By accurately measuring the mass-to-charge ratio (m/z) of proteins or peptides, mass spectrometry (MS) enables comprehensive characterization of proteins in complex samples, including:
1. Protein Identification:
Rapidly and accurately identifies proteins in samples by matching mass spectrometry data with protein databases. Additionally, MS based proteomics analysis can detect amino acid sequence information of proteins.
2. Protein Quantification:
Measures relative or absolute protein abundance using labeling methods (e.g., TMT, iTRAQ) or label-free quantification (LFQ).
3. Post-Translational Modifications (PTMs) Analysis:
Precisely detects modifications such as phosphorylation, glycosylation, and acetylation, and identifies their modification sites.
4. Protein Interaction Studies:
Reveals protein interaction networks by combining mass spectrometry with techniques such as co-immunoprecipitation (Co-IP) or affinity purification.
Leveraging the advanced mass spectrometry platform, MtoZ Biolabs offers a comprehensive range of MS based proteomics analysis service including protein identification, quantitative proteomics, protein sequencing, and post-translational modifications analysis. We provide a complete suite of proteomics solutions tailored to your research needs.
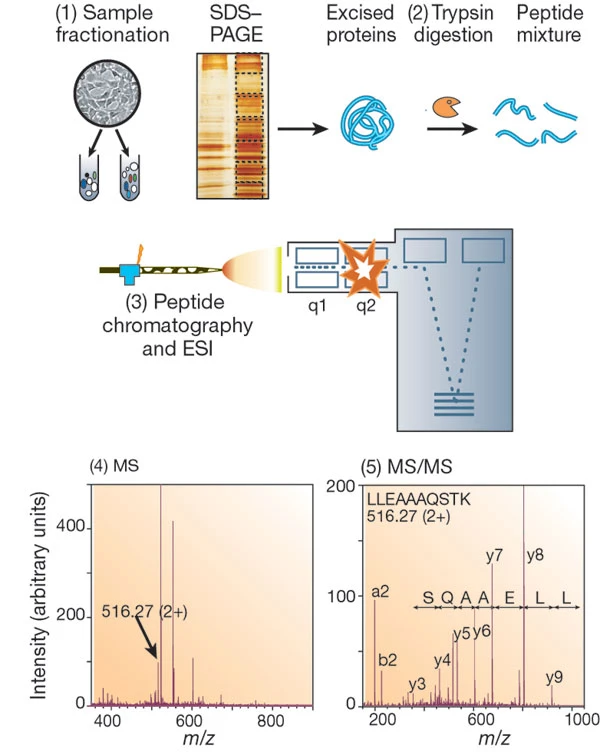
Aebersold R. et al. Nature. 2003.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced MS based proteomics analysis service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all MS based proteomics analysis data, providing clients with a comprehensive data report.
Sample Submission Suggestions
Sample Types
We accept a wide variety of sample types, including but not limited to cells, tissues, and serum/plasma.
Sample Amounts
Cell Samples: ≥1×10⁷ cells;
Tissue Samples: ≥50 mg;
Liquid Samples: ≥50 µL;
Specific sample amounts can be customized based on project requirements.
FAQ
Q. How to Validate Mass Spectrometry Results?
Validation of mass spectrometry results is a critical step in ensuring data credibility and biological significance. Below are common methods and strategies for result validation:
1. Technical Validation
Targeted Mass Spectrometry Validation: Use PRM (Parallel Reaction Monitoring) or SRM (Selected Reaction Monitoring) to verify target proteins or peptides with high sensitivity to ensure the accuracy of detection results.
Reproducibility Tests: Perform independent technical and biological replicates to validate result consistency.
2. Biological Validation
Western Blot Analysis: Use antibodies to validate protein expression levels and modification states.
Co-Immunoprecipitation (Co-IP): In protein interaction studies, use Co-IP to confirm specific binding partners of target proteins.
Functional Assays: Design knockout, overexpression, or point mutation experiments to validate the role of target proteins or modifications in biological processes.
3. Data Validation
Database Comparison: Cross-check mass spectrometry data with known databases (e.g., UniProt, PhosphoSitePlus) to confirm whether identified proteins or modifications have established biological significance.
Statistical Validation: Apply rigorous statistical methods (e.g., FDR < 1%) to filter data, ensuring the significance and accuracy of the results.
4. Integrated Validation
Multi-Omics Integration: Combine mass spectrometry results with transcriptomics, metabolomics, or other omics data to validate the biological consistency of target proteins.
Clinical Sample Validation: In disease-related studies, use additional clinical samples to validate biomarkers or targets and ensure their broader applicability.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study conducted a comprehensive proteomic analysis of patient serum samples using MS based proteomics to identify and quantify differential proteins associated with the early pathological processes of type 2 diabetes mellitus (T2DM). The results revealed that these potential biomarkers are involved in key biological pathways such as metabolic regulation and inflammatory responses, providing novel molecular insights and technical support for the early diagnosis and treatment of T2DM.
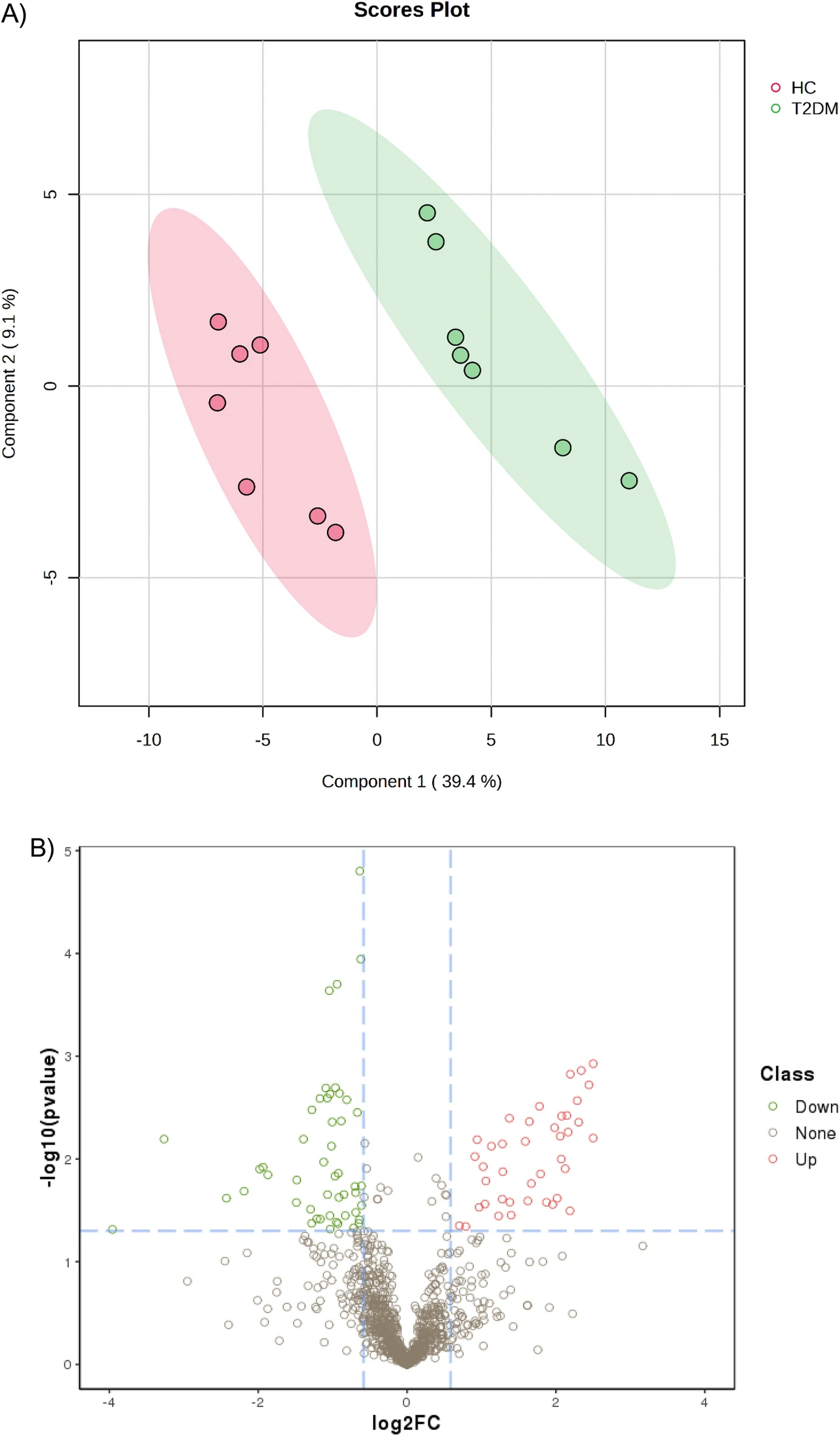
Nimer R M. et al. Sci Rep. 2023.
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MS-Based Post-Translational Modification Analysis Service
How to order?







