Mass Spectrometry Based Proteomics Analysis Service
Mass spectrometry based proteomics analysis service integrates mass spectrometry platforms with bioinformatics tools to provide a comprehensive perspective on the types, abundance, dynamic changes, and functional networks of proteins in biological samples. Unlike traditional single target protein analysis, mass spectrometry based proteomics emphasizes a holistic analysis of complex systems. Whether constructing protein expression profiles, precisely mapping post-translational modifications, or dynamically capturing protein-protein interaction networks, mass spectrometry offers comprehensive and high-resolution data support. Leveraging liquid chromatography-tandem mass spectrometry (LC-MS/MS) technology, MtoZ Biolabs provides mass spectrometry based proteomics analysis service including but not limited to qualitative and quantitative proteome analysis, post-translational modification proteomics analysis, and protein interaction network construction.
Analysis Workflow
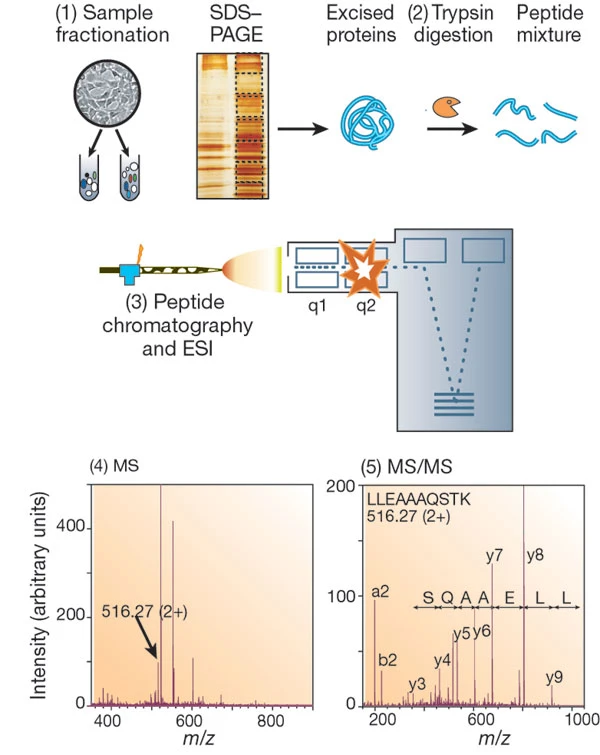
Aebersold R. et al. Nature. 2003.
Sample Preparation
Extract protein samples, followed by purification and concentration to ensure sample quality. Digest the protein samples to generate peptides for subsequent analysis.
Liquid Chromatography Separation
Separate peptides using nano-liquid chromatography to reduce sample complexity.
Mass Spectrometry Detection
Collect data with a high-resolution mass spectrometer, generating peptide mass spectra and secondary fragmentation spectra.
Data Analysis and Reporting
Identify proteins and their post-translational modifications through database searches and bioinformatics tools. Calculate relative or absolute protein abundances. Deliver a detailed experimental report, including a protein identification list, abundance changes, modification information, and biological function annotations.
Applications
Basic Research
Mass spectrometry based proteomics analysis service supports protein function analysis, post-translational modification research, and structural biology, etc. For example, it can be used to study protein interactions and construct protein interaction networks.
Disease Research
This service aids in deciphering disease mechanisms, discovering biomarkers, and supporting personalized medicine. For example, differential proteomics can help identify potential biomarkers for disease diagnosis, prognosis, and treatment response.
Drug Development
Mass spectrometry based proteomics analysis service can be used for target discovery and verification, drug mechanism research and drug safety evaluation, etc. For example, functional proteomics can screen disease-associated protein targets.
Biotechnology and Industrial Applications
It supports quality control of bioproducts, food and environmental research, and synthetic biology, etc. For example, it can detect protein components in food, analyze allergens or functional proteins.
FAQ
Q. How to Design an Experimental Plan to Ensure the Achievement of Research Objectives?
1. Define Research Objectives
Targeted Research: If the goal is to study specific proteins or modifications, choose targeted mass spectrometry (PRM/SRM) or a modification-specific enrichment strategy.
Global Exploration: For whole proteomics studies, choose labeled quantification (e.g., TMT/iTRAQ) or label-free quantification (LFQ).
2. Sample Type and Preparation
Sample Selection: Choose appropriate sample sources (e.g., cells, tissues, serum) based on research needs.
Sample Treatment: Use optimized protein extraction methods to ensure high purity and minimal degradation. Apply specific preprocessing steps (e.g., desalting, enrichment) depending on the target.
3. Quantification Strategy Selection
High-Throughput: Use multiple labeling quantification (e.g., TMT) to increase sample processing throughput.
Complex Samples: LFQ is suitable for complex samples and avoids biases introduced during labeling.
4. Biological Replicates and Controls
Replicates: Include at least three biological replicates to enhance statistical reliability.
Controls: Incorporate negative controls, positive controls, or background samples to distinguish specific results from non-specific signals.
5. Optimization of Technical Parameters
Liquid Chromatography Separation: Choose HPLC or Nano-LC systems based on sample complexity, and optimize gradient conditions.
Mass Spectrometry Mode: Select appropriate fragmentation modes (e.g., HCD, ETD) and acquisition modes (e.g., DIA or DDA).
6. Bioinformatics Integration
Combine functional annotation (e.g., GO, KEGG) and network analysis (e.g., STRING, Cytoscape) to design subsequent data processing workflows, ensuring that results are closely aligned with biological questions.
By clearly defining objectives and carefully designing the experimental plan, the scientificity and feasibility of the research plan can be ensured, maximizing the acquisition of high-quality data.
Case Study
This study utilized mass spectrometry based proteomics analysis to comprehensively analyze the protein expression profiles of esophageal cancer patients. Multiple key molecular biomarkers and post-translational modifications associated with cancer progression were identified. Based on these data, this study defined molecular subtypes of esophageal cancer and uncovered the specific biological characteristics and potential therapeutic targets of each subtype, providing a critical foundation for personalized treatment strategies for esophageal cancer.
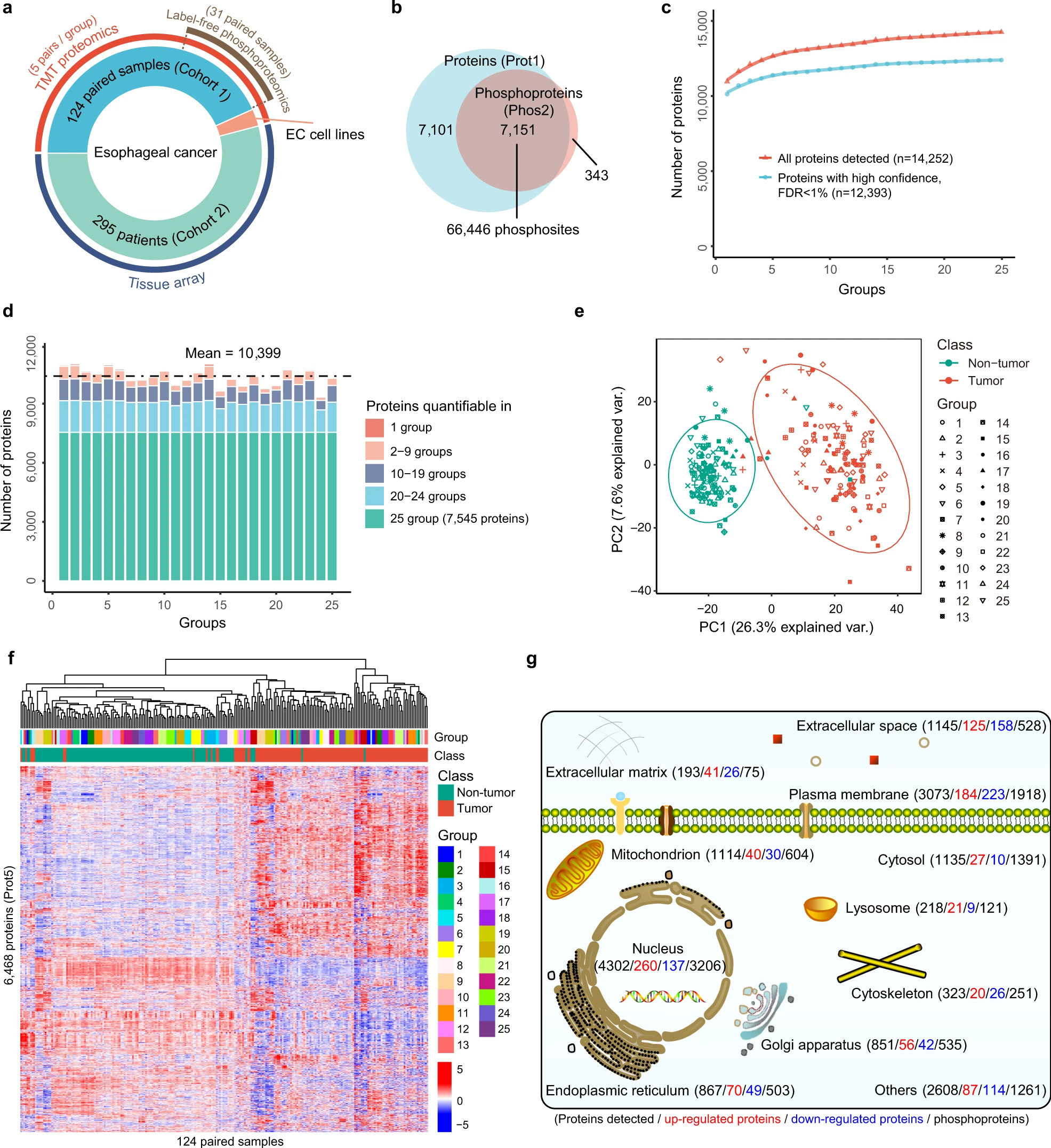
Liu W. et al. Nat Commun. 2021.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MS-Based Post-Translational Modification Analysis Service
How to order?







