Lipoylation Proteomics Service
Lipoylation proteomics focuses on investigating the mechanisms underlying protein Lipoylation modifications and their biological functions. Lipoylation refers to the covalent attachment of fatty acids (such as palmitic acid or myristic acid) to specific amino acid residues (such as cysteine or glycine) in proteins through thioester or amide bonds, thereby regulating protein membrane localization, stability, and signal transduction. The core principle of this service involves enriching lipidated proteins, identifying modification sites and types using high-resolution mass spectrometry, and applying bioinformatics analysis to elucidate their biological significance.
Lipoylation proteomics service is widely applied across various fields. In disease mechanism research, they are used to analyze aberrant palmitoylation-driven pathway activation in cancer or lipidated protein aggregation in Alzheimer’s disease. In metabolic disorders, they help uncover abnormal modifications of insulin signaling proteins in diabetes. In immune regulation, they facilitate the study of T cell receptor Lipoylation in mediating immune responses. In pathogen infection, they are employed to explore how viruses exploit host Lipoylation machinery for entry. These services also provide critical data for the development of drugs targeting Lipoylation enzymes and support precision medicine initiatives.
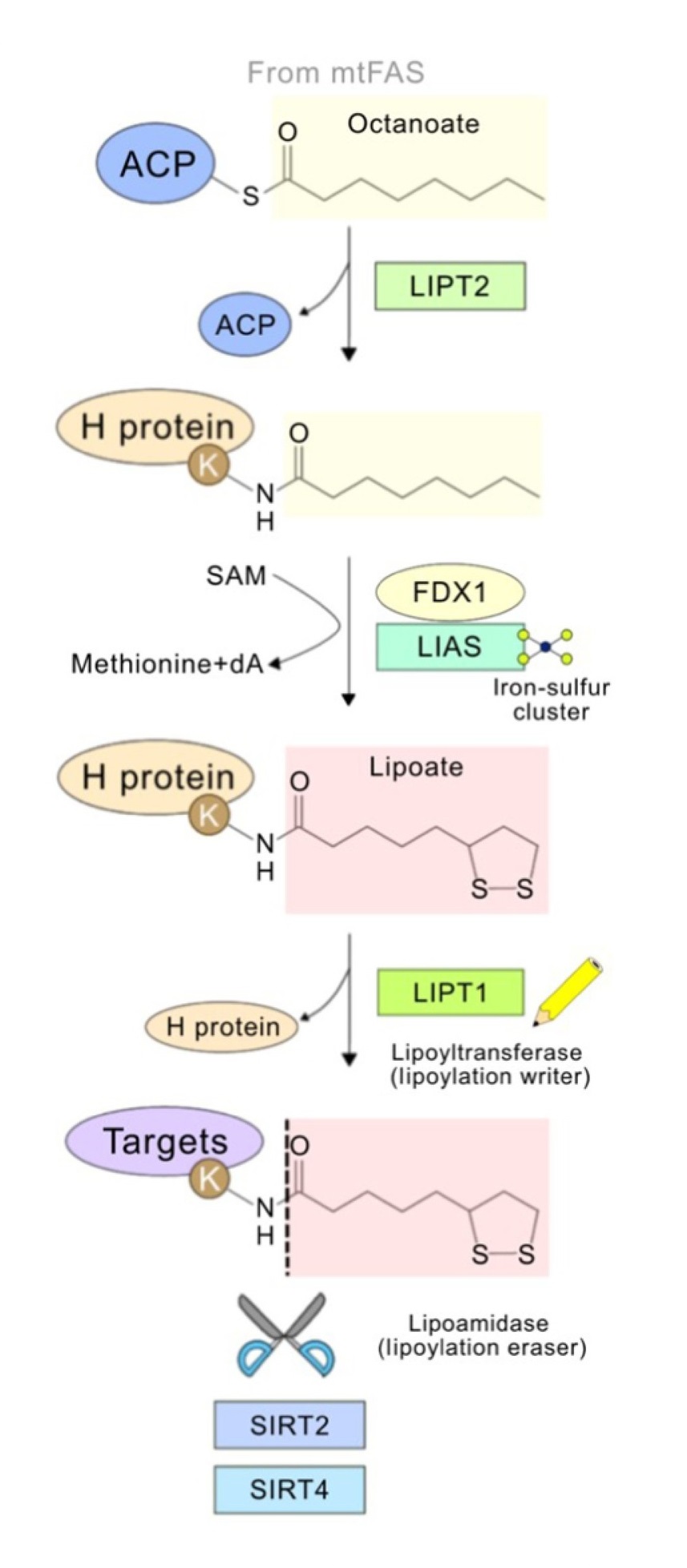
Lin, C H. et al. Trends in Biochemical Science, 2024.
Figure 1. The General Pathway of Protein Lipoylation.
Services at MtoZ Biolabs
Based on advanced instrumentation and analytical technologies, MtoZ Biolabs has launched a lipoylation proteomics service that enables systematic analysis of lipoylation types (such as palmitoylation and myristoylation), modification sites, and modification levels in protein samples. This service utilizes chemical enrichment in combination with mass spectrometry to accurately identify lipoylation sites formed through thioester or amide bonds, and generates quantitative data (such as modification abundance differences). Additionally, it provides bioinformatics annotation (including functional pathways and disease associations). The resulting data help elucidate the molecular mechanisms of lipoylation in disease, metabolism, or signaling regulation, offering valuable insights for target discovery and translational research. Common analytical methods include the following:
1. Mass Spectrometry (LC-MS/MS)
Utilizes high-resolution liquid chromatography–tandem mass spectrometry to identify and localize lipoylation sites on proteins. This method provides peptide sequences, modification positions, and relative abundance, and is the most sensitive and widely used approach for lipoylation detection.
2. SDS-PAGE (SDS–Polyacrylamide Gel Electrophoresis)
Used to separate protein samples, typically followed by Western blot with anti-lipoylation antibodies. It allows preliminary assessment of the presence of lipoylation modifications and the molecular weight range of modified proteins.
3. Immunoprecipitation (IP)
Employs lipoylation-specific antibodies to enrich target lipoylated proteins or peptides, enhancing detection of low-abundance modifications. This method is commonly used in combination with mass spectrometry or Western blotting.
4. Western Blot
Applies lipoylation-specific antibodies to detect whether a target protein is lipoylated. This technique is suitable for validation studies or targeted detection of specific proteins.
Analysis Workflow
1. Protein Extraction and Digestion
Target proteins are extracted from cells, tissues, or purified protein samples, followed by reduction and alkylation treatments. Proteins are then digested using trypsin or other enzymes to preserve lipoylation modifications and generate peptides suitable for mass spectrometry analysis.
2. Enrichment of Modified Peptides
Selective enrichment of lipoylated peptides is performed using anti-lipoylation-specific antibodies or chemical derivatization strategies, enhancing the detection of low-abundance modifications and reducing background interference.
3. Mass Spectrometry Detection and Analysis
The enriched peptides are analyzed using high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS) platforms to acquire information on lipoylation sites, peptide sequences, and modification abundance.
4. Data Interpretation and Result Output
Database searching and specific algorithms are applied to identify lipoylation sites. High-confidence spectra, modification sites, and quantitative data are reported to support downstream functional studies and mechanistic exploration.
Sample Submission Suggestions
1. Sample Types
A wide range of sample sources is supported, including cells, tissues, purified proteins, and cell lysates. It is recommended to use samples with high expression levels of mitochondria-associated proteins to enhance lipoylation modification detection efficiency.
2. Buffer Requirements
Avoid using buffers containing high salt, SDS, glycerol, EDTA, or other components that may interfere with mass spectrometry analysis or compromise the stability of lipoylation modifications.
3. Sample Storage and Transport
Samples should be stored at –80°C and shipped on dry ice. Repeated freeze-thaw cycles should be avoided to prevent loss of modifications or protein degradation. For liquid samples, concentration or lyophilization is recommended to improve sample stability.
Service Advantages
1. High-Sensitivity Detection
Powered by high-resolution mass spectrometry platforms such as Orbitrap and optimized sample preparation workflows, this service enables precise identification of low-abundance lipoylation sites, making it ideal for complex sample analysis.
2. Multi-Strategy Validation
Combines mass spectrometry, Western blot, and immunoprecipitation techniques to enhance the specificity and reliability of modification identification, supporting both mechanistic and functional studies.
3. Tailored Enrichment Protocols
Offers customized lipoylated peptide enrichment strategies that significantly improve detection rates, particularly suited for studies involving mitochondrial proteins or metabolism-related pathways.
4. End-to-End Technical Support
Delivers a complete workflow from sample processing and targeted enrichment to data analysis and bioinformatic annotation, empowering researchers to systematically investigate the roles of lipoylation in physiological and pathological contexts.
Applications
1. Metabolic Regulation Studies
Lipoylation is extensively involved in key metabolic pathways such as glycolysis and the citric acid cycle. This service can be used to elucidate its regulatory roles in energy metabolism and nutrient sensing.
2. Disease Mechanism Investigation
Lipoylation proteomics service enables the analysis of lipoylation dynamics in pathological conditions such as neurodegenerative diseases, metabolic disorders, and cancer, aiding in the identification of potential therapeutic targets.
3. Mitochondrial Function Research
Lipoylation primarily occurs on specific enzyme complexes within mitochondria, making it suitable for studying its role in the respiratory chain, oxidative stress response, and mitochondrial homeostasis.
4. Drug Response and Target Validation
Lipoylation proteomics service can be applied to evaluate dynamic changes in lipoylation modifications before and after drug treatment, supporting pathway validation and the discovery of novel drug targets.
Case Study
1. Quantitative Site-Specific Chemoproteomic Profiling of Protein Lipoylation
This study aims to develop a chemoproteomic strategy for site-specific quantitative analysis of protein lipoylation, systematically profiling the distribution and function of this modification in cells. Using human cell lines as a model, the researchers employed a chemical probe to specifically label lipoylation sites, followed by biotin-based affinity enrichment and high-resolution LC-MS/MS for detection, with stable isotope labeling for quantification. Multiple high-confidence lipoylation sites were identified, predominantly located on mitochondrial metabolism-related enzymes, and exhibited dynamic changes under different metabolic conditions. The study demonstrates that this method offers high throughput and high site-resolution advantages, providing an effective tool to explore the role of lipoylation in metabolic regulation and disease mechanisms.
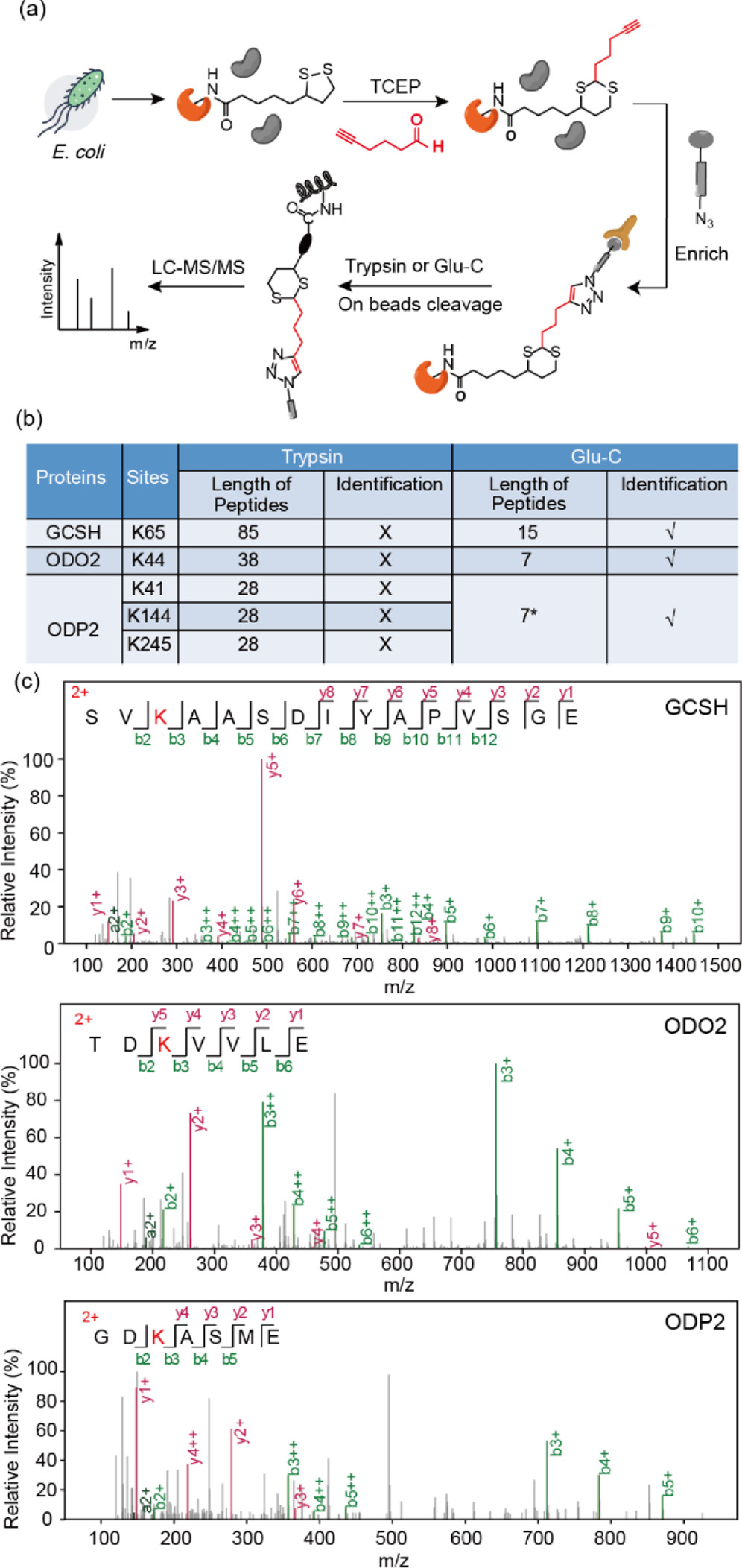
Lai, S C. et al. Journal of the American Chemical Society, 2022.
Figure 2. Identification of Lipoylation Sites in Proteomes of E. coli.
FAQ
Q1: Do you Support the Detection of Low-Abundance Lipoylated Peptides?
A1: Yes. We utilize high-sensitivity mass spectrometry platforms combined with specific enrichment strategies, such as anti-lipoylated lysine antibody enrichment, to effectively detect low-abundance lipoylated peptides even in complex sample backgrounds.
Q2: Can you Compare Samples under Different Treatment Conditions?
A2: Yes. We offer both label-free and TMT-based quantification strategies according to client needs, enabling comparative analysis of lipoylation levels across different experimental groups, which is ideal for pharmacodynamic studies and pathway screening.
How to order?







