Label-Free PTM Quantitative Analysis Service
Label-free PTM quantitative analysis is a technique that enables the relative quantification of post-translational modifications (PTMs) in proteins without the need for stable isotope labeling or chemical tags, relying instead on signal intensities derived from mass spectrometry. The method is based on high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS), where changes in modification levels are assessed by comparing the peak areas or ion intensities of modified peptides across different samples. It is suitable for studying dynamic modifications such as phosphorylation, acetylation, methylation, and ubiquitination.
The label-free PTM quantitative analysis service is widely used in the study of signal transduction pathways, biomarker discovery, drug mechanism investigation, and the regulation of cell cycle and stress responses. It is particularly well-suited for integrative multi-omics analysis and for quantification in the absence of reference standards. With advantages such as high throughput, label-free operation, and relatively low cost, label-free quantification holds significant value in both basic and translational medical research.
Services at MtoZ Biolabs
Based on advanced high-resolution mass spectrometry platforms and optimized sample preparation and data analysis workflows, the label-free PTM quantitative analysis service offered by MtoZ Biolabs enables highly sensitive identification and quantification of post-translational modifications in proteins. This service is suitable for complex samples such as cells and tissues, delivering high-quality data including modification sites, peptide sequences, spectral information, and relative quantification. It supports researchers in gaining deep insights into the functional roles of protein modifications in physiological and pathological processes.
Analysis Workflow
1. Protein Extraction and Digestion
Total proteins are extracted from samples such as cells or tissues, followed by reduction, alkylation, and trypsin digestion to ensure the preservation of modified peptide segments.
2. Enrichment of Modified Peptides
Specific enrichment strategies (e.g., phosphorylation- or acetylation-specific methods) are employed to improve the detection rate of target modified peptides.
3. LC-MS/MS Analysis
High-resolution mass spectrometry platforms (e.g., Orbitrap) are used to perform liquid chromatography–tandem mass spectrometry, capturing signal intensity data of peptide segments.
4. Data Analysis and Quantitative Comparison
Specialized software normalizes and statistically analyzes peptide ion intensities across samples, delivering results on modification sites, relative abundance, and variation trends, supporting functional annotation and downstream mechanistic studies.
Sample Submission Suggestions
1. Sample Types
A wide range of sample sources is supported, including cells, tissues, serum/plasma, and purified proteins. It is recommended that sample origin be clearly defined and processing be consistent to enhance the accuracy and comparability of quantitative analysis.
2. Buffer System
Avoid using buffers containing SDS, high salt, glycerol, or other components that may interfere with mass spectrometry detection. If specific buffers are used, please inform us in advance so that compatibility can be assessed.
3. Sample Transport
Samples should be stored at –80°C and shipped on dry ice to maintain the stability of protein modifications and prevent degradation due to freeze-thaw cycles. Sample preservation and handling guidance is available upon request.
Service Advantages
1. No Labeling Required, Streamlined Workflow
Does not rely on stable isotopes or chemical tags, offering a simplified and efficient experimental process suitable for large-scale PTM quantification studies.
2. High-Throughput Quantification
Utilizes high-resolution mass spectrometry platforms to detect multiple types of post-translational modifications simultaneously, enabling comprehensive evaluation of modification levels.
3. High Sensitivity and Reproducibility
Combines optimized peptide enrichment and data processing workflows to accurately detect low-abundance modifications with consistent and reliable results.
4. End-to-End Service
Covers the entire workflow from sample preparation and PTM enrichment to mass spectrometry detection and data analysis, supporting mechanistic research and PTM regulatory network interpretation.
Applications
1. Signal Pathway Regulation Studies
Label-free PTM quantitative analysis service enables quantification of modification level changes in key signaling molecules such as kinase substrates, uncovering the regulatory roles of PTMs in signal transduction.
2. Drug Mechanism Evaluation
By analyzing protein modification changes before and after drug treatment, potential drug targets and responsive pathways can be identified, supporting drug development and target validation.
3. Disease Mechanism Exploration
Label-free PTM quantitative analysis service is applicable to fields such as cancer, autoimmune disorders, and neurodegenerative diseases, facilitating the investigation of abnormal PTM patterns associated with disease progression.
4. Multi-Omics Integration
Can be integrated with transcriptomic, metabolomic, or other datasets to construct multilayer regulatory networks, enhancing the depth and systematic interpretation of biological discoveries.
Case Study
1. Quantitative Label-Free Phosphoproteomics Reveals Differentially Regulated Protein Phosphorylation Involved in West Nile Virus-Induced Host Inflammatory Response
This study aimed to systematically investigate the host cell phosphorylation regulatory mechanisms triggered by West Nile virus (WNV) infection using label-free quantitative phosphoproteomics. Human U251 glioma cells were used as a model, and LC-MS/MS analysis identified 1,657 phosphorylated proteins, with 853 showing significant changes upon infection. Functional enrichment analysis revealed that these differential proteins were mainly involved in inflammation-related pathways such as MAPK, NF-κB, and mTOR. Further validation using siRNA knockdown of GSK3β, PNKP, and RB1 demonstrated their negative regulatory effects on p65 activity and inflammatory cytokine expression. The findings indicate that WNV infection induces a rewiring of specific phosphorylation networks and suggest that the associated proteins may serve as potential targets for anti-inflammatory or antiviral intervention.
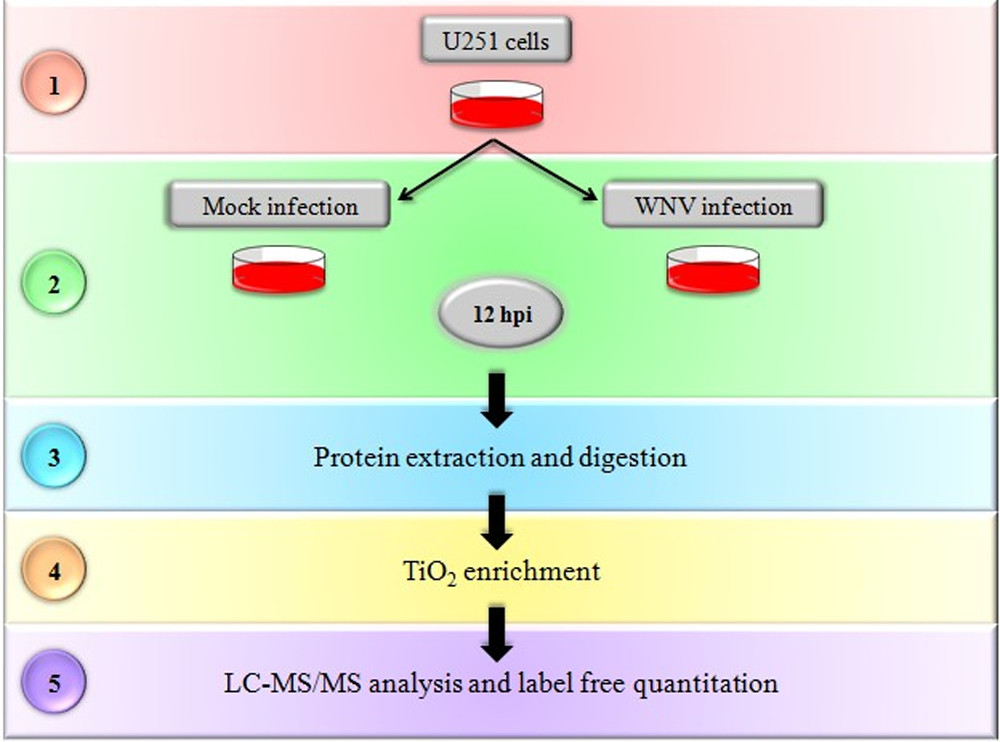
Zhang, H. et al. Journal of Proteome Research, 2015.
Figure 1. Workflow of Label-Free, Quantitative LC–MS/MS Phosphoproteomics.
2. Ubiquitination Plays an Important Role during the Formation of Chicken Primordial Germ Cells
This study aimed to elucidate the regulatory role of ubiquitination during the formation of chicken primordial germ cells (PGCs) using label-free quantitative ubiquitin proteomics. Using the differentiation process from chicken embryonic stem cells to PGCs as a model, ubiquitin-modified lysine residues were enriched and analyzed via LC-MS/MS, identifying 724 upregulated and 138 downregulated ubiquitination sites. Functional enrichment analysis revealed that these differentially modified proteins are involved in key signaling pathways, including cell cycle regulation, Wnt, MAPK, and insulin pathways. Further investigation showed that dysregulation of NEDD8 modification disrupts normal PGC formation. The results demonstrate that ubiquitination plays a central role in PGC development and provide valuable insights into the molecular mechanisms underlying avian germ cell differentiation.

Gong, W. et al. Journal of Animal Science, 2024.
Figure 2. Overview of the Ubiquitomic Analysis.
FAQ
Q1: What Types of Post-Translational Modifications (PTMs) Can this Service Detect?
A1: This service can detect a wide range of common PTMs, including but not limited to phosphorylation, acetylation, methylation, ubiquitination, and glycosylation. Specific enrichment strategies can be customized according to project requirements.
Q2: Can Low-Abundance Modifications Be Detected?
A2: Yes. We utilize high-sensitivity mass spectrometry platforms (e.g., Orbitrap) combined with optimized enrichment methods, enabling the detection of low-abundance modified peptides, even in complex biological samples.
Q3: Does the Service Support Quantitative Comparison?
A3: Yes. By comparing ion intensities of modified peptides across experimental groups, the service provides relative quantification of modification sites to reveal dynamic changes under different treatment conditions.
How to order?







