How to Use Protein Sequence Analysis for Functional Prediction?
-
High-quality protein sequence analysis
-
Cross-validation and annotation using multiple databases
-
Structural prediction, molecular docking, and integrated pathway analysis
-
Functional prediction reports tailored for candidate drug targets
With the rapid advancement of high-throughput sequencing technologies, an increasing number of novel proteins have been identified. However, a significant proportion of these proteins remain functionally unannotated. In this context, protein sequence analysis serves as an essential initial step toward understanding protein function. This review systematically summarizes widely adopted strategies for protein sequence analysis and discusses how the integration of multiple bioinformatics tools can enhance the accuracy and reliability of functional prediction.
How Does Protein Sequence Reveal Function?
The amino acid sequence of a protein determines its three-dimensional structure, which in turn governs its function. While protein sequences can vary greatly across species, conserved domains and functional motifs are frequently preserved and serve as key indicators for functional inference. For example, ATP-binding sites and phosphorylation sites in kinase proteins are often highly conserved at the sequence level. Additionally, the principle of functional conservation among homologous proteins underpins the strategy of homology-based functional inference.
Core Strategies for Functional Prediction Based on Protein Sequence
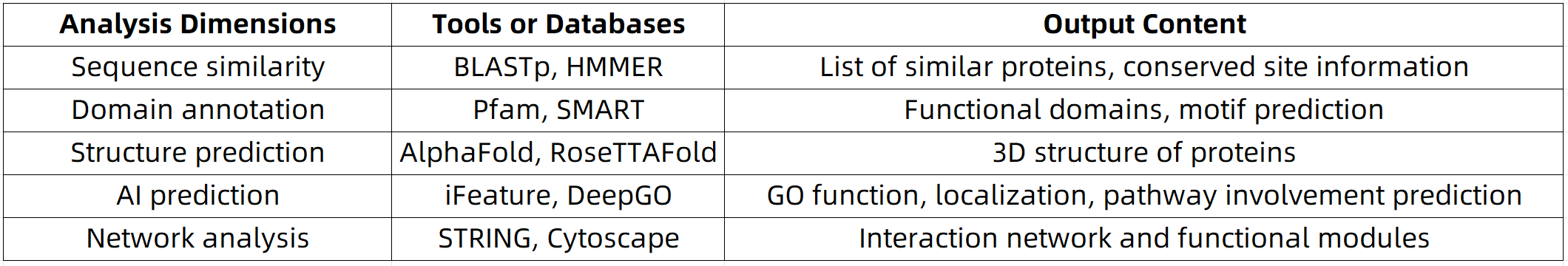
1. Homology Search
Identifying proteins with similar sequences in annotated databases is one of the most straightforward and effective methods for inferring protein function. Common tools include:
(1) BLASTp: A widely used tool for rapid local or online sequence alignment. It is recommended to use an E-value threshold of <1e-5 to ensure the reliability of the results.
(2) HMMER: Based on Hidden Markov Models (HMMs), this tool offers high sensitivity for detecting conserved domains and is frequently used for domain annotation with the Pfam database.
2. Domain Identification and Functional Annotation
Protein domains represent structural and functional units. By detecting known domains within a protein sequence, one can infer the protein’s potential functional roles. Databases such as Pfam, SMART, and InterProScan aggregate extensive domain annotations and are commonly employed for this purpose.
3. Protein Structure Prediction and Functional Inference
Recent breakthroughs in deep learning, particularly with models like AlphaFold2, have made it possible to predict protein tertiary structures with high confidence, even in the absence of experimental data. Structural predictions facilitate:
(1) Identification of catalytic sites and ligand-binding pockets
(2) Structural alignment with proteins of known function
(3) Construction of protein–ligand docking models for activity prediction
4. Sequence-Based Feature Extraction and Machine Learning-Based Prediction
In cases where no close homologs are available, machine learning offers a promising alternative. Features such as amino acid composition, secondary structure probabilities, and physicochemical properties can be extracted from sequences and used to train predictive models.
(1) Common features: Amino acid composition (AAC), dipeptide composition (DPC), position-specific scoring matrix (PSSM) profiles
(2) Typical algorithms: Support Vector Machine (SVM), Random Forest, Convolutional Neural Networks (CNN), Transformer models
(3) Applications: Predicting Gene Ontology (GO) terms, subcellular localization, and protein–protein interactions
5. Integration of Protein–Protein Interaction (PPI) Networks and Functional Modules
Proteins usually operate within complex interaction networks rather than functioning independently. By analyzing interaction partners from PPI databases such as STRING and BioGRID, one can infer the functional context of a target protein at the systems level.
(1) If the target protein interacts with multiple key regulators within known signaling pathways, it likely participates in the corresponding biological processes
(2) Incorporating Graph Neural Networks (GNNs) can further refine prediction accuracy by modeling the topological structure of the interaction network
Integrating Multiple Strategies to Enhance the Reliability of Functional Prediction
Predictions based on a single strategy are often biased; therefore, it is advisable to adopt an integrated approach combining multiple methods to improve prediction accuracy. For instance, Graph Neural Networks (GNNs) can be utilized to integrate heterogeneous data sources and refine functional inference.
How Can MtoZ Biolabs Accelerate Your Functional Prediction Research?
At MtoZ Biolabs, we recognize the critical importance of protein function annotation in both fundamental research and drug discovery. Leveraging our proteomics and metabolomics platforms, we offer a comprehensive suite of services integrating AI-driven prediction, structural modeling, and function annotation, including:
Our expert team is proficient in mainstream bioinformatics tools and can provide customized analyses based on specific project requirements. Whether you have newly identified a potential protein or require high-throughput functional screening, we offer robust, data-driven support for comprehensive function annotation.
Protein function prediction is a complex yet highly promising endeavor. Starting from sequence information, it is possible to elucidate a protein’s biological roles by integrating approaches such as homology modeling, structural prediction, AI inference, and network analysis. With MtoZ Biolabs’ one-stop functional annotation solution, you can accelerate your research and uncover deeper insights into the molecular mechanisms of life.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







