How to Use Affinity Purification-Mass Spectrometry (AP-MS) for Protein–Protein Interaction (PPI) Detection?
- Mapping of tumor signaling pathway interaction networks
- Identification of complexes involving epigenetic modification enzymes
- Screening of drug–target interaction candidates
- Network analysis of stem cell fate regulation
High-throughput protein–protein interaction (PPI) detection represents a crucial approach for systematically elucidating cellular signaling pathways, deciphering protein functions, and understanding the architecture of biological networks. Among various PPI detection methods, Affinity Purification–Mass Spectrometry (AP-MS) is widely recognized as one of the most reliable and physiologically relevant techniques. It has been extensively applied across multiple biological systems, including mammalian cells, model organisms, and disease models.
Overview of AP-MS: Principles and Advantages
1. Principle of AP-MS
AP-MS is an affinity-based proteomic approach that relies on antibody recognition or tag-mediated purification to isolate a target (bait) protein along with its interacting partners (prey) from complex biological mixtures. The purified complexes are then subjected to mass spectrometry for PPI detection.
Typical workflow:
(1) Expression of a tagged bait protein (e.g., Flag, HA, Strep, BioTag)
(2) Cell lysis followed by affinity purification to capture protein complexes associated with the bait
(3) Elution of the protein complexes and enzymatic digestion (typically using trypsin)
(4) LC–MS/MS analysis for peptide and protein identification
(5) Bioinformatic processing to remove nonspecific interactors and construct high-confidence interaction networks
2. Advantages of AP-MS over Traditional Methods
(1) High Specificity: Affinity purification using specific antibodies or tags effectively minimizes background noise.
(2) High Throughput: Integration with automated platforms and mass spectrometry enables parallel processing of hundreds of samples.
(3) Physiological Relevance: Protein complexes are assembled in living cells, reducing artificial artifacts and false positives.
(4) Quantitative Capability: Both label-based (e.g., TMT, iTRAQ) and label-free (LFQ) quantitative strategies can be employed to assess interaction dynamics.
Designing an Efficient AP-MS PPI Detection Experiment
1. Selection of Bait Protein and Tagging Strategy
The choice of bait protein should account for factors such as expression level, structural properties, and subcellular localization. Tag selection is also critical, as it influences both purification efficiency and complex integrity.
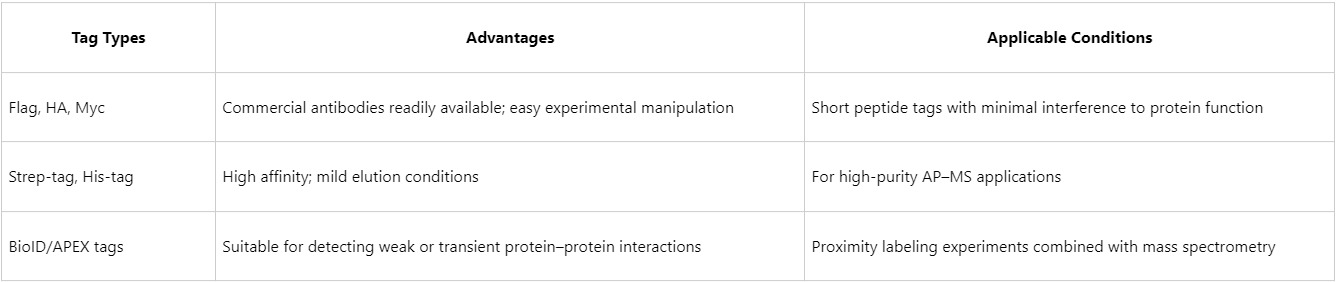
2. Experimental System and Protein Expression Methods
(1) For low-abundance endogenous proteins, overexpression systems combined with affinity tags are recommended.
(2) For endogenous interaction studies, CRISPR-mediated knock-in tagging is preferred to maintain native regulation.
(3) The choice of cell model (e.g., HEK293, HeLa, tumor-derived, or stem cell lines) should align with the biological context of interest.
3. Optimization of Purification Conditions
Efficient affinity purification aims to preserve genuine interactions while minimizing nonspecific binding. Critical parameters include:
(1) Buffer composition: Physiological salt concentrations and detergent types (e.g., NP-40, Triton X-100) influence complex stability.
(2) Elution methods: Options include competitive, acidic, or enzymatic elution, each with distinct effects on complex integrity.
Mass Spectrometry Strategies: Qualitative, Quantitative, and Filtering
1. LC–MS/MS Platform Selection
(1) High-resolution mass spectrometers such as the Orbitrap Exploris 480 or QE HF-X are well suited for complex proteomic analyses.
(2) NanoLC–MS systems are recommended to enhance detection sensitivity, particularly for low-abundance PPI detection.
2. Quantitative Approaches: LFQ vs. TMT
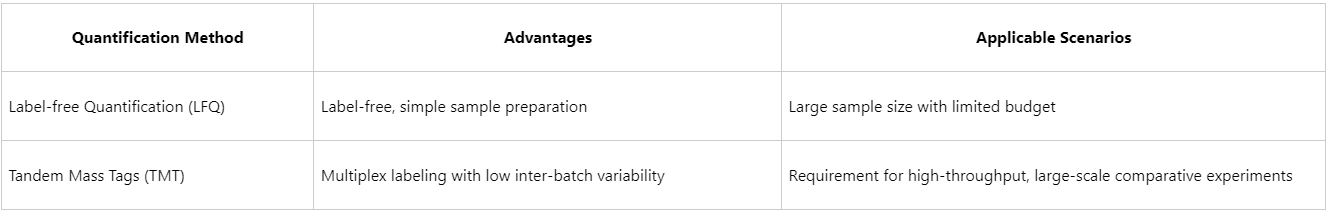
3. Distinguishing True Interactions from Nonspecific Backgrounds
(1) Include appropriate control samples (e.g., empty vector or unrelated protein).
(2) Apply computational algorithms such as SAINT, MiST, or CompPASS to estimate interaction confidence scores.
(3) Cross-reference results with the CRAPome database to exclude known contaminants.
(4) Validate reproducibility using biological replicates to confirm stable interactions.
Data Interpretation and Network Construction
1. Functional Enrichment and Network Visualization
(1) Perform GO and KEGG pathway enrichment analyses to infer biological functions of the identified complexes.
(2) Visualize PPI networks using Cytoscape or similar tools to identify key hub proteins.
(3) Integrate multi-omics datasets (e.g., transcriptomic or phosphoproteomic data) to gain deeper mechanistic insights.
2. Validation of Key Interactions
(1) Confirm core interactions using co-immunoprecipitation (Co-IP) and Western blotting.
(2) Assess functional outcomes of interaction disruption via CRISPR-mediated knockdown or knockout experiments.
(3) Employ pharmacological perturbation or mutant assays to evaluate the biological significance of specific interactions.
Technical Strengths of MtoZ Biolabs in AP-MS
As a leading omics research service provider, MtoZ Biolabs has completed hundreds of AP-MS projects in PPI detection encompassing:
We offer a comprehensive workflow covering experimental design, protein expression, affinity purification, mass spectrometry, and bioinformatic analysis, enabling efficient and reliable protein interaction research.
High-throughput AP-MS–based PPI detection requires meticulous optimization at every stage, from experimental design to data interpretation. By integrating rigorous methodologies with advanced instrumentation, researchers can obtain high-confidence PPI detection maps that elucidate the dynamic regulatory mechanisms underlying biological processes. MtoZ Biolabs provides professional expertise and a robust experimental platform to facilitate the exploration of protein interaction networks, signaling pathways, and potential therapeutic targets in diverse research contexts.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







