How to Use Acetylation Data for Pathway Enrichment Analysis in Research?
-
Chromatin-associated proteins (e.g., histones), influencing gene expression
-
Metabolic enzymes (e.g., GAPDH, CS, IDH), regulating catalytic activity
-
Signal transduction molecules, modulating protein–protein interactions and subcellular localization
-
Facilitates the construction of metabolic and signaling pathway networks.
-
Provides clear visualization of enrichment results, with support for protein position annotation tracking.
-
Accessible via tools such as the R package clusterProfiler, DAVID, and KOBAS.
-
Enables functional interpretation from three domains: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC).
-
Incorporates enrichment scores to evaluate the magnitude of regulatory effects.
-
The interplay between acetylation and other PTMs, such as phosphorylation
-
Organelle-specific pathway enrichment (e.g., mitochondria versus nucleus)
-
Reconstruction of disease-associated modification networks
Acetylation is a critical protein post-translational modification (PTM) that plays essential roles in diverse biological processes, including the regulation of cellular metabolism, signal transduction, and chromatin remodeling. With the advancement of high-resolution mass spectrometry technologies, researchers can systematically detect and identify acetylation sites, thereby enabling in-depth investigation of their biological functions. Within the vast datasets generated from acetylation studies, pathway enrichment analysis serves as a powerful approach for elucidating the underlying regulatory mechanisms at the systems level. Conducting such analysis requires not only accurate quantification and differential assessment of acetylation sites, but also the mapping of modified proteins to relevant biological pathways, followed by the application of multiple databases and computational algorithms to uncover potential regulatory networks. Through enrichment analysis, researchers can characterize modification patterns within key pathways and infer their functional alterations under specific physiological or pathological conditions. This article provides a systematic overview, from a scientific perspective, of methodologies for performing pathway enrichment analysis based on acetylation data, and illustrates, with practical tools and case examples, how to extract meaningful biological insights from modification proteomics datasets.
Functional Background of Acetylation Modification
Protein acetylation primarily comprises N-terminal acetylation and lysine acetylation, with the latter being notably reversible and subject to precise regulation by acetyltransferases (e.g., p300/CBP) and deacetylases (e.g., HDACs, SIRTs).
This modification is widely distributed among:
Consequently, large-scale acetyl-proteomics datasets have become an invaluable resource for investigating epigenetic regulation and elucidating disease mechanisms.
Basic Workflow of Pathway Enrichment Analysis
1. Data Preparation: Identification and Quantification of Acetylated Proteins
High-resolution mass spectrometry platforms (e.g., Orbitrap Fusion, QE HF-X), when combined with anti-acetyl-lysine enrichment strategies, enable the detection of thousands of acetylation sites alongside their quantitative measurements (e.g., TMT labeling, label-free quantification). Comprehensive acetyl-proteomics services, covering sample preparation, modification enrichment, and mass spectrometry identification, can ensure both high proteome coverage and robust data quality.
2. Differential Analysis: Identification of Biologically Relevant Modification Changes
(1) Select significantly altered acetylation sites (e.g., log2FC > 1 and p < 0.05)
(2) Map acetylation sites to their corresponding proteins to compile a list of differentially acetylated proteins
3. Pathway Enrichment Analysis
Widely used databases and tools include:
(1) KEGG (Kyoto Encyclopedia of Genes and Genomes)
(2) GO (Gene Ontology)
(3) Reactome, WikiPathways, and other supplementary databases can further enhance pathway coverage and interpretive depth.
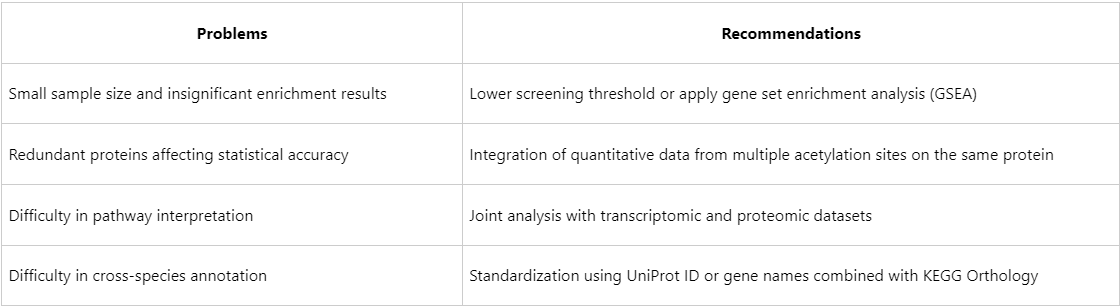
Common Issues and Analysis Recommendations for Acetylation Data
Summary and Outlook
Pathway enrichment analysis of acetylation datasets not only facilitates the understanding of protein functional regulation, but also reveals their roles at the systems level in both physiological and pathological contexts. Looking ahead, the integration of multi-omics datasets will enable further exploration of:
In all cases, high-quality acetyl-proteomics data form the foundation for reliable analysis. Leveraging advanced mass spectrometry platforms and stringent quality control procedures, comprehensive proteomics service providers, MtoZ Biolabs, can offer robust technical support to researchers, thereby accelerating scientific discovery.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







