How to Select an Appropriate PPI Detection Technique?
- Principle: The proteins of interest are fused separately to the DNA-binding and activation domains of a transcription factor; interaction between the two reconstitutes transcriptional activity and activates reporter gene expression.
- Advantages: High throughput, technically simple, suitable for identifying novel interactions.
- Limitations: High false-positive rate, restricted to nuclear proteins, unsuitable for detecting interactions involving membrane or transmembrane proteins.
- Principle: A fluorescent protein is split into two fragments, each fused to a protein of interest; interaction between the proteins restores fluorescence.
- Advantages: Enables visualization and subcellular localization, suitable for membrane proteins
- Limitations: Irreversible complementation and signal accumulation complicate quantification.
- Principle: Specific antibodies enrich a target protein together with its interacting partners, which are subsequently identified by Western blotting or mass spectrometry.
- Advantages: High specificity, well-suited for validating established interactions.
- Limitations: Requires high-quality antibodies. Weak or transient interactions are easily lost.
- Principle: A protein fused with a GST tag is immobilized on beads to capture interacting partners, which are subsequently detected after elution.
- Advantages: Appropriate for validating recombinant protein interactions in vitro.
- Limitations: Non-physiological conditions, not reflective of the intracellular environment.
- Principle: Protein complexes are enriched using affinity tags or antibodies and analyzed by LC-MS/MS to identify interaction partners.
- Advantages: High throughput, quantitative, applicable at endogenous expression levels.
- Limitations: Complex background, requires optimized purification conditions.
- Principle: The protein of interest is fused to a promiscuous biotin ligase, which labels proximal proteins with biotin in living cells; labeled proteins are subsequently identified by LC-MS.
- Advantages: Captures weak and transient interactions, suitable for membrane proteins.
- Limitations: Requires fusion protein expression. Labeling radius may extend beyond true interactors.
- Principle: Crosslinking reagents covalently stabilize protein complexes, which are then digested and analyzed by mass spectrometry to identify crosslinked peptides and infer interaction sites.
- Advantages: Provides spatial information on interaction interfaces, valuable for structural studies.
- Limitations: Challenging data analysis, limited crosslinking efficiency.
- Extensive expertise in membrane protein and signaling pathway research
- Support for multiple approaches, including BioID, AP-MS, and XL-MS
- Reproducible, low-background immunoprecipitation protocols
- Dedicated bioinformatics support for interaction map construction and visualization
Protein–Protein Interactions (PPIs) form the foundation of most cellular processes. From signal transduction and transcriptional regulation to metabolic networks, PPI networks illuminate the dynamic regulatory mechanisms underlying protein functions and biological pathways. Accurate detection and characterization of PPIs are therefore of paramount importance in elucidating disease mechanisms, identifying novel therapeutic targets, and facilitating drug development. A wide range of PPI detection methods is available, spanning from yeast two-hybrid assays to state-of-the-art high-throughput mass spectrometry platforms, each with distinct strengths and limitations. This review provides a systematic overview of the principles and applications of mainstream PPI detection techniques, aiming to aid researchers in selecting the most appropriate strategy according to their experimental objectives.
What Is PPI Detection and Why Is It Critical?
PPI detection refers to experimental approaches designed to identify and validate physical or functional interactions between proteins. In disease research, aberrant protein interactions often indicate underlying pathogenic mechanisms; in drug discovery, PPI interfaces have emerged as promising targets for both small- and macromolecule-based therapeutics. With the advancement of systems biology and structural biology, PPIs are no longer viewed solely as binary binding events but rather as dynamic regulatory networks. Selecting an appropriate PPI detection strategy is thus a prerequisite for advancing fundamental scientific inquiries.
Mainstream PPI Detection Techniques
PPI detection techniques can be broadly divided into three categories: reporter system–based approaches, immunoprecipitation-based methods, and mass spectrometry–based high-throughput strategies.
1. Reporter System–Based Classical Approaches
(1) Yeast Two-Hybrid (Y2H)
(2) BiFC (Bimolecular Fluorescence Complementation)
2. Immunoprecipitation-Based Co-precipitation Methods
(1) Co-Immunoprecipitation (Co-IP)
(2) GST Pull-Down
3. Mass Spectrometry–Based High-Throughput Methods
(1) AP-MS (Affinity Purification coupled with Mass Spectrometry)
(2) BioID / TurboID
(3) Cross-linking MS (XL-MS)
Criteria for Selecting a PPI Detection Technique
The choice of method depends on the research objective, sample source, and type of interaction under investigation. Preliminary guidance is summarized in Table X.
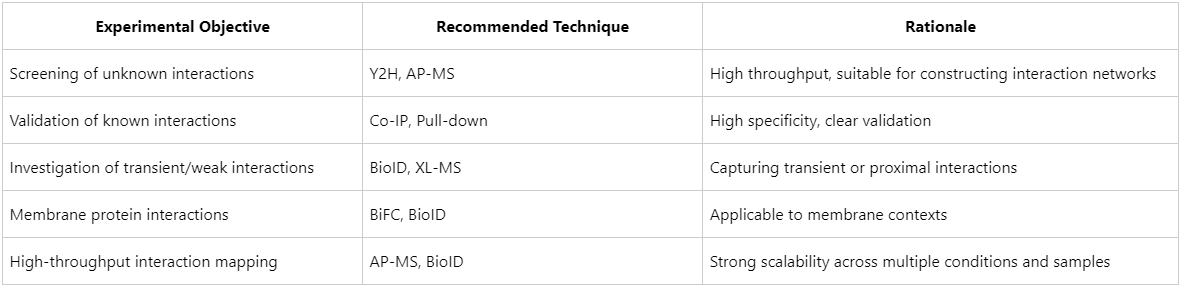
In practice, the optimal strategy often involves combining multiple techniques to balance sensitivity, specificity, and spatial resolution. For instance, AP-MS can be used to identify candidate interaction partners, which may then be validated using Co-IP, followed by XL-MS to pinpoint interaction interfaces.
MtoZ Biolabs: An Integrated Platform for PPI Research
PPI studies frequently require complex sample preparation and high-precision analytical platforms. Leveraging advanced Orbitrap Exploris high-resolution mass spectrometry, MtoZ Biolabs offers comprehensive services spanning interaction partner identification, network reconstruction, and interaction site mapping.
Key advantages include:
PPIs represent an indispensable tool in modern life sciences, with the critical challenge being the alignment of detection strategies with the scientific question at hand. While no single method can address all research needs, careful consideration of experimental objectives, sample constraints, and methodological limitations allows researchers to select strategies that maximize both efficiency and discovery potential. Professional support and advanced mass spectrometry platforms, such as those provided by MtoZ Biolabs, can further assist in generating high-quality protein interaction maps.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







