How to Improve Sensitivity and Specificity in Acetylated Protein Quantification: Techniques and Reagents
-
Employing lysis buffers containing HDAC inhibitors (e.g., Trichostatin A, Nicotinamide)
-
Conducting rapid lysis at low temperatures to minimize reaction duration
-
Optimizing protein concentration and total yield to prevent dilution that could hinder detection of modified proteins
Acetylation, a pivotal post-translational modification (PTM), plays a critical role in chromatin remodeling, metabolic regulation, and signal transduction. With the continuous advancement of mass spectrometry-based proteomics, investigations into acetylated proteomes have deepened considerably. However, owing to the inherently low abundance, transient nature, and dynamic regulation of acetylation, improving the sensitivity and specificity of acetylated protein quantification analyses remains a central challenge for researchers.
Sample Preparation: Protein Extraction and Inhibition of Deacetylation
Acetylation modifications are highly susceptible to the activity of endogenous enzymes following cell lysis. Thus, effective inhibition of histone deacetylase (HDAC) activity during sample preparation is essential. Recommended measures include:
Furthermore, avoiding surfactants such as SDS, which can interfere with antibody binding, can enhance downstream enrichment efficiency.
Enrichment of Modified Peptides: Key Determinants of Detection Sensitivity
Given that acetylated peptides constitute a minute fraction of the total peptide pool, highly efficient and specific enrichment is a prerequisite for sensitive detection. Common strategies include:
1. Affinity Enrichment Using Anti-Acetyl-Lysine Antibodies
(1) Affinity purification with anti-acetyl-lysine (anti-Kac) monoclonal antibodies is currently the most widely adopted approach.
(2) Monoclonal antibodies coupled to Protein A/G magnetic beads are preferred over traditional agarose beads due to superior binding efficiency and reproducibility.
(3) Antibody sources should be selected from commercially available or custom-developed products validated by mass spectrometry.
2. Chemical Derivatization and Specific Affinity Approaches (Exploratory)
(1) Amino-affinity materials in combination with isothiocyanate derivatization can selectively target peptides bearing acetyl groups.
(2) These methods remain at an exploratory stage and are particularly suited for in-depth characterization of specific modification sites.
Strategies for Acetylated Protein Quantification
Analysis of acetylated protein quantification generally falls into two categories: stable isotope labeling methods (e.g., TMT, iTRAQ) and label-free quantification.
1. TMT/iTRAQ Peptide Labeling
(1) Advantages: Enables multiplexed sample analysis with high quantitative precision.
(2) Challenges: Low abundance of acetylated peptides and limited labeling efficiency.
(3) Optimization: Perform antibody-based enrichment prior to labeling and remove unreacted peptides to improve both efficiency and reproducibility.
2. Label-Free Quantification
(1) Suitable for studies with limited sample availability or budget constraints.
(2) When combined with high-resolution mass spectrometry platforms (e.g., Orbitrap Exploris 480) and precise alignment algorithms, this approach can achieve quantitative consistency comparable to isotope-labeling methods.
Mass Spectrometry Platforms and Data Analysis: Instrumentation as a Determinant of Quantitative Depth
1. Importance of High-Resolution Mass Spectrometry
(1) Instruments such as Orbitrap Fusion Lumos and the Exploris series offer high sensitivity and dynamic range, making them ideal for acetylated peptide analysis.
(2) Employing higher-energy collisional dissociation (HCD) can enhance the identification efficiency of b/y fragment ions.
2. Data Processing Strategies
(1) Utilize dedicated PTM quantification software such as MaxQuant or Proteome Discoverer.
(2) Appropriately configure parameters such as FDR thresholds, peptide length, and acetylation site localization probability (Localization Probability > 0.75).
(3) Incorporate machine learning-based feature filtering and functional enrichment analyses to maximize biological interpretability.
Summary and MtoZ Biolabs's Integrated Solution
The sensitivity and specificity of acetylated protein quantification rely on the coordinated optimization of every step, from sample preparation through to mass spectrometry analysis. key improvement strategies:
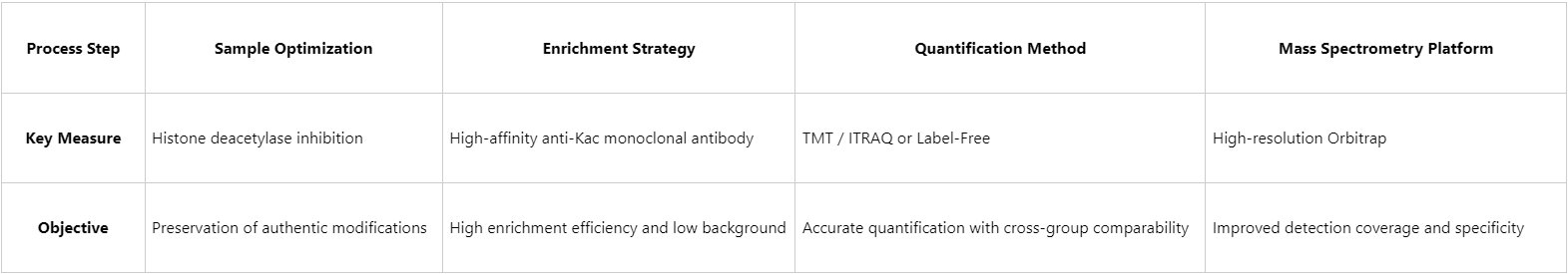
Acetylation is among the most prevalent PTMs in eukaryotes, playing central roles in transcriptional regulation, metabolism, and signal transduction pathways. The rapid development of proteomics technologies, particularly mass spectrometry platforms, has enabled systematic elucidation of the roles of protein acetylation in diverse physiological and pathological processes. Leveraging over a decade of expertise in proteomics research, MtoZ Biolabs has established a comprehensive acetylated protein quantification platform, providing end-to-end services from sample preprocessing to bioinformatics analysis. This platform is widely applied in research areas including tumor metabolism, epigenetic regulation, and signal transduction.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







