How to Improve Reliability and Reproducibility of PPI Experimental Results?
- Inconsistent protein expression levels: Overexpression of exogenous proteins may induce non-physiological interactions, whereas insufficient expression often results in weak or undetectable signals.
- Variability in sample processing: Subtle differences in lysis conditions, centrifugation parameters, or protein storage methods can substantially affect the stability of interaction complexes.
- High levels of non-specific binding: Particularly in affinity-based assays (e.g., co-immunoprecipitation), background signals frequently obscure genuine protein interactions.
- Methodological limitations: Different PPI detection approaches, such as Y2H and AP-MS, capture distinct interaction landscapes, resulting in limited overlap and low cross-method validation rates.
- Lack of standardized data analysis criteria: The absence of unified scoring systems or threshold definitions leads to considerable variability in analytical outcomes across laboratories.
Protein-protein interactions (PPIs) play fundamental roles in cellular signaling, metabolic regulation, and disease pathogenesis. With the rapid advancement of omics technologies, PPI research has become a central focus in life science studies. However, due to the intrinsic complexity of experimental systems and pervasive non-specific interactions, PPI experiments frequently suffer from poor reproducibility and elevated false-positive rates. Enhancing experimental reliability and reproducibility has therefore become a critical prerequisite for the accurate elucidation of protein interaction networks.
Why Is the Reproducibility of PPI Experiments Often Poor?
Prior to discussing optimization strategies, it is essential to understand the major factors contributing to poor reproducibility in PPI experiments:
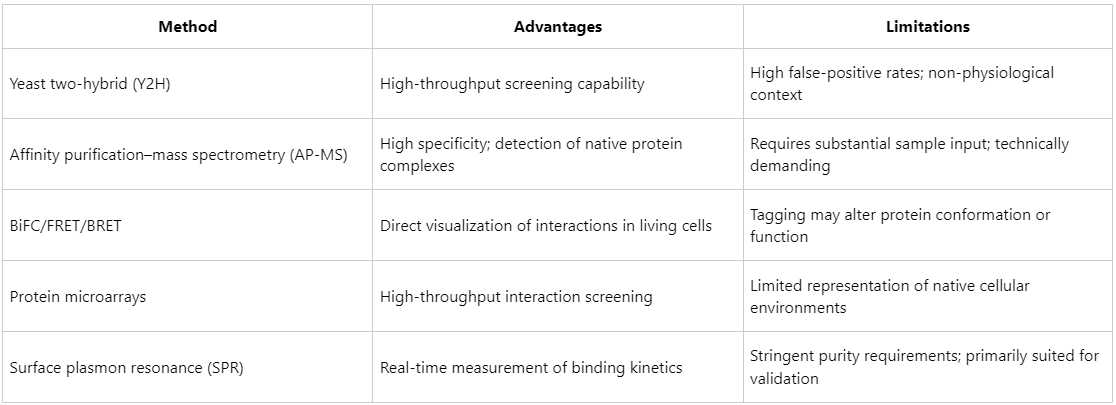
Overview and Challenges of Commonly Used PPI Detection Methods
Practical Strategies to Enhance the Reliability and Reproducibility of PPI Experiments
1. Optimization of Experimental Design
(1) Selection of appropriate tags and expression systems: Small, minimally disruptive tags (e.g., FLAG, HA, Strep) should be prioritized to reduce structural perturbations. Expression systems approximating endogenous protein levels (e.g., BAC-based systems) better reflect physiological conditions.
(2) Implementation of comprehensive positive and negative controls: Controls such as empty vectors, tag-only expression, and known interacting or non-interacting protein pairs are essential for identifying non-specific binding events.
(3) Parallel biological and technical replication: A minimum of three independent biological replicates, each processed separately, is critical for ensuring statistical robustness and reproducibility.
2. Strict Control of Sample Handling Procedures
(1) Standardization of lysis buffers and conditions: Mild lysis conditions compatible with interaction preservation (e.g., NP-40 or CHAPS) should be used, while harsh detergents such as SDS should be avoided.
(2) Maintenance of cold-chain conditions and protease inhibition: All experimental procedures should be performed at 4°C, with protease inhibitors included to prevent degradation of protein complexes.
(3) Minimization of non-specific binding: Background signals can be reduced through pre-clearing steps, the use of high-affinity antibodies or agarose beads, and careful optimization of washing and elution conditions.
3. Integration of Multi-Omics Validation Strategies
(1) Statistical filtering of mass spectrometry data: Algorithms such as SAINT, MiST, or CompPASS should be employed to quantitatively assess interaction confidence, rather than relying solely on binary detection.
(2) Contextual filtering using transcriptomic or proteomic data: Interactions involving proteins with extremely low or absent expression in the same cellular context are more likely to represent false positives.
(3) Cross-platform validation: Key interactions should be corroborated using orthogonal approaches, such as combining AP-MS with BiFC or Y2H, to enhance overall confidence.
Recommendations for Data Analysis and Visualization
1. Adoption of Standardized Data Processing Pipelines
A recommended workflow includes data cleaning, quantitative analysis, background subtraction, interaction scoring, and network construction. PPI networks can be visualized using Cytoscape, with GO and KEGG enrichment analyses applied to identify critical regulatory modules.
2. Establishment of a Unified Interaction Confidence Scoring Framework
Results should be cross-referenced with established databases such as BioGRID, STRING, and IntAct. Customized confidence criteria (e.g., detection in more than three independent replicates combined with a SAINT score > 0.9) can substantially improve analytical consistency.
As a central approach for elucidating signaling pathways and disease mechanisms, the reliability of protein interaction studies directly determines the validity of downstream biological interpretations. Through rigorous experimental design, standardized workflows, and the integration of high-resolution mass spectrometry platforms with multi-dimensional validation strategies, both the reproducibility and credibility of PPI experiments can be markedly enhanced. MtoZ Biolabs is dedicated to providing high-confidence PPI research solutions, encompassing the entire workflow from sample preparation and affinity purification to mass spectrometry analysis and data interpretation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







