How to Choose the Right Peptide Sequencing Method? Edman Degradation vs. Mass Spectrometry
-
Protein identification of known sequences: Mass spectrometry is preferred due to its speed and rich information content.
-
Analysis of unknown protein sequences: De novo sequencing by mass spectrometry is the first choice, with Edman degradation providing complementary verification of the N-terminal sequence when required.
-
N-terminal sequence confirmation: Edman degradation enables direct and unambiguous reading of the N-terminal, making it suitable for validating the initiation sites of antibodies or recombinant proteins.
-
Post-translational modification mapping: Mass spectrometry offers powerful capabilities in pinpointing modification sites, supporting studies of phosphorylation, acetylation, and other regulatory modifications.
-
Complex samples with high-throughput demands: Mass spectrometry provides superior efficiency and broad adaptability.
-
Single or repeated verification needs: Edman degradation yields stable and reproducible sequence data, making it valuable for structural confirmation.
Peptide sequencing is an essential approach for elucidating the primary structure of proteins, and it has been widely applied in protein identification, validation of novel sequences, localization of modification sites, and antibody development. Currently, Edman degradation and mass spectrometry (MS) represent the two mainstream peptide sequencing methods, each offering distinct advantages tailored to different experimental requirements. This article provides a systematic comparison of these methods in terms of principles, performance, and applicability, aiming to assist researchers in selecting the most appropriate sequencing strategy.
Technical Principles
1. Edman Degradation: Sequential Identification via Chemical Cleavage
In Edman degradation, phenyl isothiocyanate (PITC) reacts with the N-terminal amino acid of a peptide chain, enabling the stepwise removal and identification of each N-terminal residue to sequentially read the peptide sequence. This linear sequencing method yields unambiguous results and operates independently of database searches.
2. Mass Spectrometry: Mass-to-Charge Analysis of Ion Fragments
Mass spectrometry-based sequencing, typically performed using tandem mass spectrometry (MS/MS), involves ionizing peptide fragments and inducing their fragmentation. The resulting mass-to-charge (m/z) spectra are then analyzed to reconstruct the amino acid sequence. When coupled with database searches or de novo algorithms, this method is applicable to both known and novel peptides.
Comparison of Technical Performance
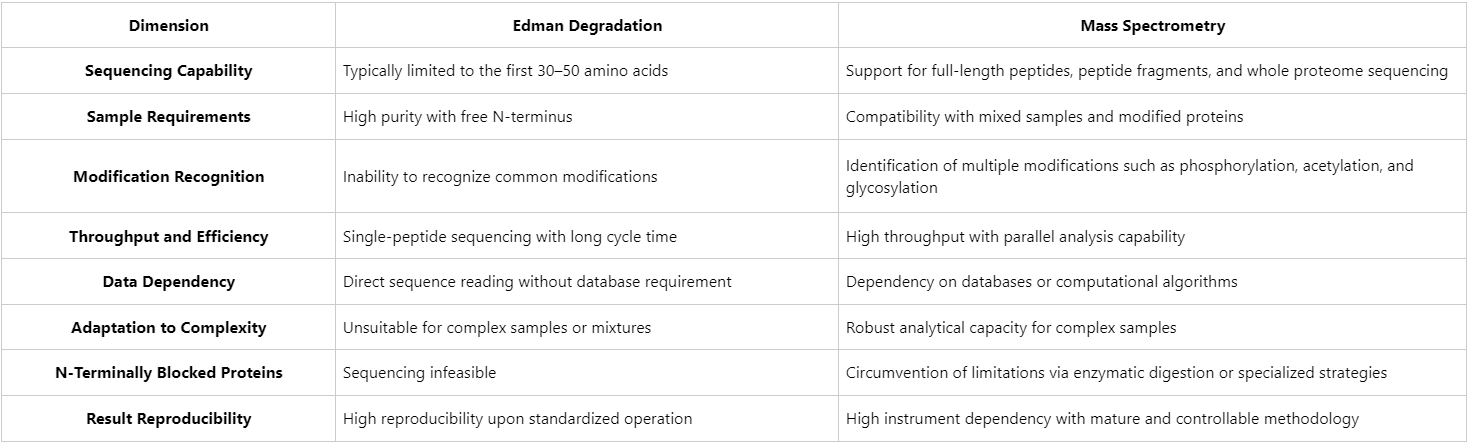
Figure1. Edman Degradation vs. Mass Spectrometry
Recommendations for Application Scenarios
The choice of peptide sequencing method can be guided by research objectives and sample characteristics as follows:
Development Trends and Integrated Applications
With continuous advances in instrument resolution, fragmentation algorithms, and database modeling, mass spectrometry has become the predominant tool in proteomics and functional protein research. Its high throughput and multidimensional analytical capacity ensure broad applicability in both basic research and biopharmaceutical development. Nevertheless, Edman degradation remains indispensable in scenarios requiring exceptional precision, particularly for direct N-terminal validation, eliminating algorithmic uncertainties, or confirming the correct expression of engineered proteins. In practice, researchers can combine Edman degradation with mass spectrometry to achieve more comprehensive and reliable peptide structural information.
MtoZ Biolabs offers customized peptide sequencing methods tailored to specific sample types and research objectives, balancing efficiency and accuracy, and supporting diverse applications from basic research to industrial translation. Through integrated technical solutions, we aim to help researchers obtain precise and trustworthy protein sequence data. For project consultation and technical support, please feel free to contact us.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







