How to Analyze Protein Disulfide Bonds?
- Loss of function (e.g., inactivated antibodies)
- Protein aggregation or formation of insoluble species
- Increased immunogenicity risk (commonly encountered in drug development)
- Prepare separate reduced and non-reduced samples
- Perform proteolysis (e.g., using trypsin)
- Conduct LC–MS/MS analysis
- Simple workflow applicable to a broad range of proteins
- Enables identification of peptides linked by disulfide bonds
- Does not allow precise localization of the linked cysteine residues
- Limited resolution for complex proteins (e.g., antibodies with multiple disulfide bonds)
- Perform proteolysis under non-reducing conditions
- Apply ETD or ECD fragmentation during MS/MS
- Accurately determine cross-linked peptides and cysteine positions
- Preserves disulfide bond structure in situ
- Enables accurate Cys–Cys pairing identification
- Particularly effective for structurally complex proteins (e.g., IgG)
- Requires advanced instrumentation (e.g., Orbitrap Fusion, Exploris series)
- Data interpretation is more computationally demanding
- Maintains the original protein conformation
- Suitable for integrity verification of protein therapeutics
- Applicable only to low-molecular-weight proteins (<50 kDa)
- Requires highly advanced instrumentation and is limited by sensitivity
- Advantages: Enables verification of functional impact
- Disadvantages: Time-consuming and not amenable to high-throughput screening
Rationale for Analyzing Protein Disulfide Bonds
Disulfide bonds play a crucial role in maintaining the stability of a protein’s tertiary structure, as well as influencing its biological activity and conformational integrity. Incorrect disulfide bond pairing can result in:
Accordingly, precise determination of the connectivity and positional arrangement of disulfide bonds is a critical step in elucidating protein structure–function relationships.
Methods for Analyzing Protein Disulfide Bonds
1. Comparative Mass Spectrometry under Reducing and Non-reducing Conditions
(1) Principle
Protease digestion and mass spectrometric analysis are performed on the same sample under both reducing conditions (e.g., DTT, TCEP treatment) and non-reducing conditions. By comparing peptide profiles, potential disulfide linkages can be inferred.
(2) Experimental Procedure
Identify differential peptides: peaks corresponding to cross-linked peptides in the non-reduced sample indicate potential disulfide bonds.
(3) Advantages
(4) Limitations
2. ETD/ECD Mass Spectrometry
(1) Principle
Electron Transfer Dissociation (ETD) and Electron Capture Dissociation (ECD) enable peptide backbone fragmentation while preserving disulfide bonds, allowing in situ maintenance of disulfide linkages and precise localization of cysteine residues.
(2) Experimental Procedure
(3) Advantages
(4) Limitations
3. Top-down Mass Spectrometry
(1) Principle
Performs MS/MS fragmentation directly on the intact protein without prior proteolysis, thereby retaining the complete native conformation, including disulfide bonds.
(2) Advantages
(3) Limitations
4. Site-directed Mutagenesis (Cys → Ser) for Functional Verification
This approach removes specific cysteine residues through point mutation, followed by functional assays and mass spectrometric analysis, to determine their involvement in key disulfide bonds.
Capabilities of MtoZ Biolabs in Protein Disulfide Bonds Analysis
MtoZ Biolabs offers high-resolution, high-sensitivity analytical solutions for protein disulfide bonds, supporting workflows from conformational studies to pharmaceutical quality control.
1. Technical Platform
(1) Orbitrap Exploris 480 / Fusion Lumos
(2) Fully equipped for ETD fragmentation
(3) High-coverage proteolysis strategies (e.g., combinations of trypsin, Lys-C, Glu-C)
2. Service Features
(1) Capability to identify heavy chain–light chain linkage sites in antibodies
(2) Construction of disulfide bond maps for both natural and recombinant proteins
(3) Access to dedicated technical advisors for experimental strategy design
Example Applications
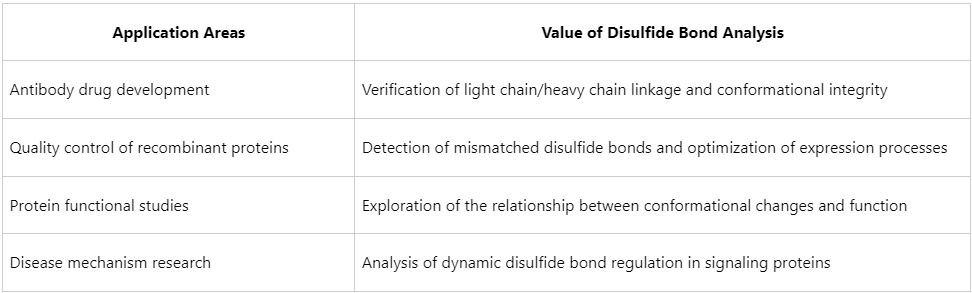
Comprehensive characterization of protein disulfide bonds facilitates a deeper understanding of structure–function relationships and holds significant value in drug development and biosynthetic research. Through the integration of advanced mass spectrometry platforms and specialized technical expertise, researchers can generate detailed structural maps of protein conformations. If you are conducting research related to protein structure, function, or drugs, please contact MtoZ Biolabs for professional disulfide bond analysis services.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







