How Does Peptidomics Bridge the Gaps in Proteomics?
- Inability to accurately identify low-abundance yet functionally critical protein cleavage fragments
- Challenges in resolving dynamic changes under complex post-translational modifications (PTMs)
- Failure to capture the biological relevance of protein degradation or processing products in vivo
Does Proteomics Truly Provide Comprehensive Coverage?
Proteomics has become a cornerstone technology for elucidating the functional states of biological systems, particularly playing a crucial role in deciphering disease mechanisms and identifying biomarkers. However, with continued advances in research, scientists have increasingly recognized that even high-throughput and highly sensitive mass spectrometry–based proteomics possesses intrinsic limitations.
These limitations include:
In this context, peptidomics has emerged as a powerful and complementary discipline to proteomics, enabling deeper exploration of disease states and molecular mechanisms from the perspective of endogenous protein fragments.
What Is Peptidomics, and How Does It Fundamentally Differ from Proteomics?
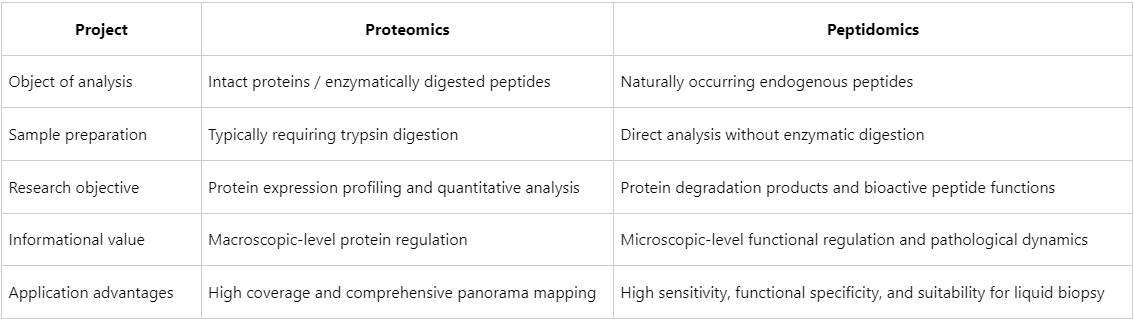
Proteomics determines which proteins are expressed, whereas peptidomics elucidates how these proteins are cleaved and processed within biological systems, and what functional roles the resulting peptides exert.
How Does Peptidomics Address the Limitations of Proteomics?
1. Missing Information on Protein Cleavage and Processing: Peptidomics Addresses the Gap in Dynamic Degradation
Many key biological processes, such as inflammation, apoptosis, and tumor metastasis, induce extensive protein cleavage and modification, changes that are often not reflected at the total protein expression level. For example, certain inflammatory signals activate metalloproteinases that cleave extracellular matrix proteins into bioactive signaling peptides. These functional fragments can be detected exclusively through peptidomic analysis, while conventional proteomics may completely overlook them. MtoZ Biolabs has developed an endogenous peptide identification database and an integrated pipeline for annotating cleavage patterns, enabling precise tracking of protein processing origins.
2. Signal Masking by High-Abundance Proteins: Peptidomics Enables Detection Beyond Conventional Limits
In biological fluids such as serum or cerebrospinal fluid, high-abundance proteins (e.g., albumin, immunoglobulin G) dominate mass spectrometry signals, often obscuring low-abundance yet biologically significant molecules. Through pre-fractionation, selective enrichment, and molecular weight filtering (typically <10 kDa), peptidomics effectively minimizes macromolecular interference, enhances trace peptide detection, and thereby improves diagnostic sensitivity.
3. Complexity of Post-Translational Modifications: Peptidomics Excels in Deciphering Localized Modifications
Proteomic analyses of PTMs, such as phosphorylation or acetylation, are often constrained by peptide redundancy and limited resolution among isomeric variants. In contrast, peptidomics directly examines naturally modified endogenous peptides, allowing more accurate characterization of regulatory mechanisms. For instance:
(1) The hydroxylation state of neurotransmitter peptides (e.g., neuropeptide Y) correlates with stress responses.
(2) The deacetylation state of certain tumor-associated signaling peptides can indicate malignancy levels.
4. Functional Molecule Identification: Peptidomics Focuses on Signal Peptides and Bioactive Fragments
Many critical signaling molecules are not full-length proteins but biologically active fragments generated through proteolytic processing, such as hormones and regulatory peptides. These short, highly specific fragments often evade detection in proteomic studies due to database or quantification limitations. Peptidomics effectively compensates for this gap by directly profiling bioactive peptides derived from protein processing.
5. Insights into Disease Heterogeneity: Peptidomics Reveals Subtle but Clinically Relevant Differences
In studies of complex and heterogeneous disorders, such as cancer and neurodegenerative diseases, global protein expression differences are frequently minimal, while subtle alterations in processing products may hold the key to pathophysiological variation. Peptidomics enables the discovery of minor yet diagnostically informative peptide changes across disease subtypes, providing valuable insights for precise molecular classification.
Peptidomics and Proteomics Integration: Synergy Beyond Standalone Analyses
Modern mass spectrometry platforms (e.g., Orbitrap Exploris, TripleTOF) now enable simultaneous acquisition of multidimensional data, making integrated proteomic–peptidomic analysis increasingly feasible.
The advantages of this integrated approach include:
(1) Offering both macroscopic (protein-level) and microscopic (cleavage product–level) perspectives
(2) Mapping the full dynamic continuum from protein expression to proteolytic processing
(3) Building pathway and network models with enhanced biological interpretability
(4) Improving biomarker robustness, specificity, and reproducibility
As a comprehensive mass spectrometry–based omics solution provider, MtoZ Biolabs offers an integrated service platform spanning proteomics, peptidomics, and metabolomics, including but not limited to:
(1) Optimized sample stratification strategies: separation of high/low molecular weight components and targeted enrichment
(2) Customized mass spectrometry workflows: combined DDA+DIA acquisition and PRM-based validation
(3) Advanced bioinformatics analysis: integrated visualization from protein to peptide level
(4) Dual support for research and translation: tailored solutions for both fundamental and industrial applications
Whether exploring novel disease mechanisms, identifying more sensitive diagnostic targets, or constructing multi-omics integrative models, MtoZ Biolabs provides professional, efficient, and reliable technical support.
Proteomics offers a comprehensive landscape of protein expression; however, the determinants of biological function and disease manifestation often reside in the finer molecular details. Peptidomics serves as the key to uncovering these subtleties, bridging the gaps that proteomics alone cannot address, and delivers deeper molecular insights for precision medicine. The integration of proteomics and peptidomics will become an indispensable omics partnership. MtoZ Biolabs continues to advance high-resolution mass spectrometry platforms and peptide recognition algorithms, empowering researchers to explore biological systems at unprecedented molecular depth and facilitating the translation from fundamental research to clinical application.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







