How Does Mass Spectrometry Reveal the Amino Acid Sequence of Proteins
-
High-resolution mass spectrometers (e.g., Orbitrap Exploris, Fusion Lumos) are employed to ensure accurate sequencing
-
Standardized pre-processing workflows, including automated enzymatic digestion, purification, and sample enrichment
-
Support for multiple quantification strategies (TMT, Label-Free, SILAC) and modules for modification detection
-
Comprehensive reporting for structural sequence restoration, mutation detection, and modification site localization
-
Integration with chemical proteomics, metabolomics, and related platforms to facilitate functional research
Introduction: The Core Pathway of Protein Structure Analysis
Protein function is determined by its structure, with the linear sequence of amino acids—known as the primary structure—forming the first hierarchical level. Mass spectrometry, particularly liquid chromatography-tandem mass spectrometry (LC-MS/MS), has emerged as a central technique for elucidating protein structure. Compared to traditional Edman degradation, mass spectrometry offers advantages such as high throughput, automation, and the capability to detect post-translational modifications. It is extensively applied in basic research and quality control of biopharmaceuticals.
Basic Principles of Mass Spectrometry Sequencing: From Peptides to Sequence
Mass spectrometry does not directly determine the protein sequence but infers the amino acid composition by analyzing the mass-to-charge ratios (m/z) and fragmentation patterns of peptide fragments. The key steps are:
1. Proteolysis: Proteins are enzymatically cleaved into defined peptide segments using specific proteases such as trypsin.
2. Liquid Chromatography (LC): Peptides are separated by high-performance liquid chromatography prior to mass spectrometry analysis to reduce sample complexity.
3. MS1: The precise molecular mass of intact peptide ions (precursor ions) is measured to obtain their mass/charge information.
4. MS2: Selected precursor ions undergo collision-induced dissociation (CID or HCD), generating fragment ions (e.g., b and y ions) with characteristic fragmentation patterns.
5. Sequence Reconstruction: By calculating mass differences between fragment ions, the differences between adjacent amino acids are deduced, enabling sequential assembly of the peptide sequence.
Overview of the Protein Mass Spectrometry Sequencing Workflow
Step 1: Sample Preparation
Protein samples are extracted and treated with reducing agents and alkylating reagents to reduce disulfide bonds and alkylate free cysteine residues.
Step 2: Proteolysis into Peptides
Proteins are digested with trypsin to generate specific peptide fragments, typically cleaving at the C-terminal of lysine (K) and arginine (R) residues.
Step 3: Liquid Chromatography Separation (LC)
Peptide mixtures are separated using high-performance liquid chromatography, improving resolution and signal quality.
Step 4: Mass Spectrometry Analysis (MS1+MS2)
Peptides enter the mass spectrometer, where precursor ion scanning (MS1) is performed. High-abundance precursor ions are selected for fragmentation in the collision cell, producing fragment ions whose spectra are recorded as MS2.
Step 5: Data Analysis and Sequence Identification
Spectra are analyzed and compared against protein databases using bioinformatics tools such as MaxQuant, PEAKS, and Byonic to identify peptides and determine their amino acid sequences.
Key Advantages of Mass Spectrometry in Deciphering Amino Acid Sequences
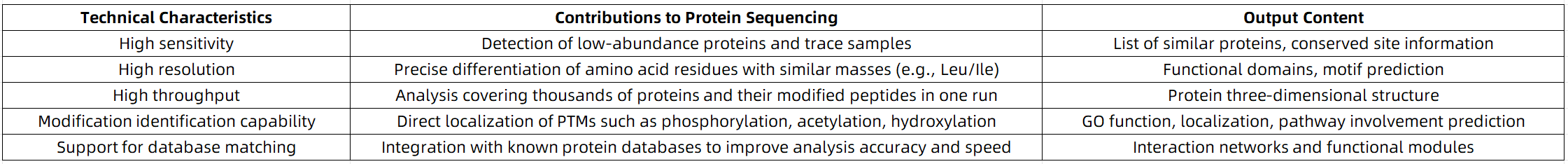
MtoZ Biolabs Protein Sequencing Services
As a leading provider of proteomics technology solutions, MtoZ Biolabs has established a comprehensive platform for primary structure protein sequencing:
Mass spectrometry, by decoding peptide fragmentation patterns, provides a robust and efficient tool for elucidating the primary structure of proteins. It not only reveals the amino acid sequence but also identifies post-translational modifications, variant sites, and splice variants, thereby offering critical data support for life sciences and biomedical research. For researchers engaged in protein structure characterization, target validation, or drug development, MtoZ Biolabs offers a high-quality, high-throughput comprehensive protein sequencing solution.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







