How Do De Novo Sequencing and Homology Search Integrate in Analysis?
In proteomics research, database-dependent search remains the dominant analytical strategy. However, its effectiveness hinges on the completeness and accuracy of reference databases. This reliance becomes a significant limitation when dealing with non-model organisms, natural products, antibody fragments, or variations at translation initiation sites, leading to potential omissions or incorrect identifications.
To address this limitation, de novo sequencing offers a database-independent approach for inferring amino acid sequences directly from MS/MS spectra. This technique is particularly advantageous for characterizing unknown peptides, structural isomers, and peptides with uncommon modifications. When integrated with homology search, it further enables downstream applications such as functional annotation, evolutionary analysis, and experimental validation—together forming a complementary and enhanced analytical framework.
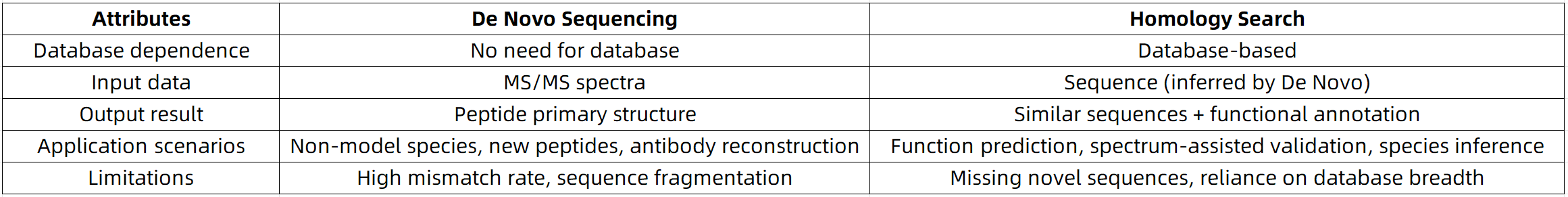
De Novo Sequencing vs. Homology Search: What Are the Fundamental Differences?
Strategies for the Integration of De Novo Sequencing and Homology Search
1. De Novo Prediction of Candidate Peptide Sequences
High-quality MS/MS spectra are subjected to de novo sequencing using tools such as PEAKS, Novor, and DeepNovo, yielding multiple high-confidence candidate peptide sequences, often comprising several isomeric variants.
2. Leveraging De Novo Results for Homology Search
The de novo-predicted peptide sequences, even if partial, can be queried against databases such as NR, SwissProt, Uniprot, or custom-built repositories using tools like BLAST, MS-BLAST, SPIDER, or the PEAKS SPIDER module to:
(1) Identify homologous proteins with significant sequence similarity;
(2) Infer functional motifs or active regions;
(3) Recover contextual sequence information flanking incomplete de novo peptide segments;
(4) Verify whether observed modifications or amino acid substitutions (e.g., R→K, M→O) represent genuine biological variations rather than technical artifacts.
3. Constructing a Composite Evidence Chain of “High-Confidence Peptide + Homologous Match”
(1) High-confidence de novo-predicted peptides provide direct sequence-level evidence;
(2) Supporting homology matches contribute functional annotation, species attribution, and structural inference;
(3) When further validated through synthetic peptide experiments or functional assays, this integrated approach enables robust and confirmatory protein identification.
Practical Applications
1. Proteomics Research on Non-Model Organisms
In studies involving species such as plants, insects, or microorganisms—where reference databases are often poorly represented—de novo sequencing serves to generate candidate peptide sequences. These can be subsequently aligned to sequences from closely related species via homology search, aiding in evolutionary tree construction and protein function network analysis.
2. Identification of Natural Bioactive Peptides or Peptides With Unknown Functions
In natural product research areas such as antimicrobial peptides, neuropeptides, and enterogastrones, de novo sequencing facilitates the discovery of novel peptide sequences. Homology-based comparison can then rapidly assess their similarity to known functional peptides, thereby expediting biological activity screening.
3. Antibody Sequence Determination and Engineering
De novo sequencing is employed to elucidate the complementarity-determining regions (CDRs) of antibody light and heavy chains. Homology alignment helps to assess their sequence similarity to known antibodies, supporting strategies such as antibody humanization and the design of Fab or single-chain variable fragments (scFv).
How to Improve the Accuracy of De Novo Sequencing and Homology Search?
1. Data-Level Optimization
(1) Employ high-resolution mass spectrometers (e.g., Orbitrap Eclipse, timsTOF Pro 2) to enhance detection sensitivity and spectral precision;
(2) Utilize multi-enzyme proteolytic digestion strategies to improve peptide coverage;
(3) Integrate advanced acquisition technologies such as PASEF or FAIMS to improve spectral quality and complexity handling.
2. Analysis-Level Optimization
(1) Compare results generated by multiple de novo sequencing algorithms to identify peptides with high inter-algorithm consistency;
(2) Incorporate post-translational modification (PTM) identification modules to reduce interference and improve interpretation accuracy;
(3) Map predicted peptide sequences back to the original spectra to reinforce confidence in identifications through a validation mechanism.
3. Homology Search-Level Optimization
(1) Leverage a flexible combination of databases, including public repositories and custom-built resources (e.g., translated transcriptomic libraries);
(2) Evaluate homologous matches across multiple species to assess sequence conservation and potential biological relevance;
(3) Employ structure-based alignment tools (e.g., HHpred) to assist in identifying functions of low-homology or divergent peptide regions.
MtoZ Biolabs provides an integrated de novo sequencing and homology search analysis workflow, offering the following features:
1. AI-Powered Peptide Prediction: Deep learning models are applied to enhance spectral interpretation accuracy and confidence in peptide identification;
2. Comprehensive Database Support: Supports homology search across multiple sources, including UniProt, NR, SwissProt, and user-defined databases;
3. Sequence Annotation and Functional Prediction: Delivers high-confidence peptide sequences with functional annotations and recommended downstream validation strategies;
4. Synthetic Peptide Validation Services: Offers synthesis and functional validation for critical peptide sequences to confirm bioactivity.
De novo sequencing enables the discovery of previously uncharacterized peptides, while homology search confers functional and biological interpretation to these sequences. When applied in tandem, these approaches not only improve the accuracy of protein identification but also provide a robust foundation for functional analysis and experimental validation. MtoZ Biolabs continuously integrates high-performance mass spectrometry platforms with advanced AI algorithms to deliver powerful and reliable solutions for unknown protein characterization. If you are exploring a combined approach to de novo sequencing and homology-based analysis, we welcome you to contact us for comprehensive technical support.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







