How Can DADA2 Be Integrated with QIIME2 for Full-Length 16S rRNA Analysis Using Third-Generation Sequencing?
To analyze full-length 16S rRNA sequencing data from third-generation platforms (e.g., PacBio, Oxford Nanopore) using both DADA2 and QIIME2, the following workflow can be applied:
1. Installation and Setup
(1) Install the latest version of QIIME2 following the official installation guidelines.
(2) DADA2 is integrated as a QIIME2 plugin and does not require separate installation.
2. Data Import
Before importing data into QIIME2 using the 'qiime tools import' command, ensure that the raw sequencing files meet the following criteria:
(1) Demultiplexed sequencing data: Each sample should have its own FASTQ file (or paired-end FASTQ files).
(2) Removal of non-biological sequences: Primers, adapters (or barcodes), and linker sequences should be removed before analysis.
(3) Paired-end sequencing consistency: If the sequencing was performed in paired-end mode, each sample should have two corresponding FASTQ files.
3. Quality Control and Denoising
(1) Denoising and quality filtering are performed using the DADA2 plugin. For PacBio or Oxford Nanopore sequencing data, the following command is commonly used:
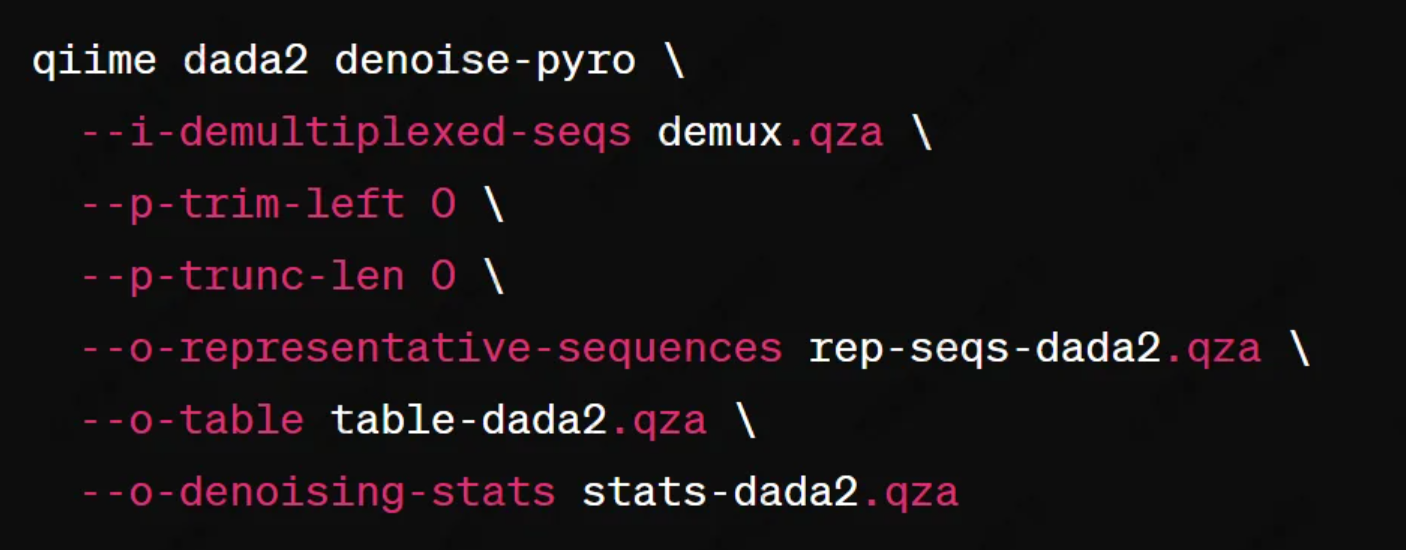
Figure 1
(2) No truncation is applied ('--p-trunc-len 0') since long-read sequencing technologies generally generate high-quality full-length reads, making length-based filtering unnecessary.
4. Biodiversity and Taxonomic Analysis
The denoised feature table (ASV table) and representative sequences are then used for downstream analyses, including:
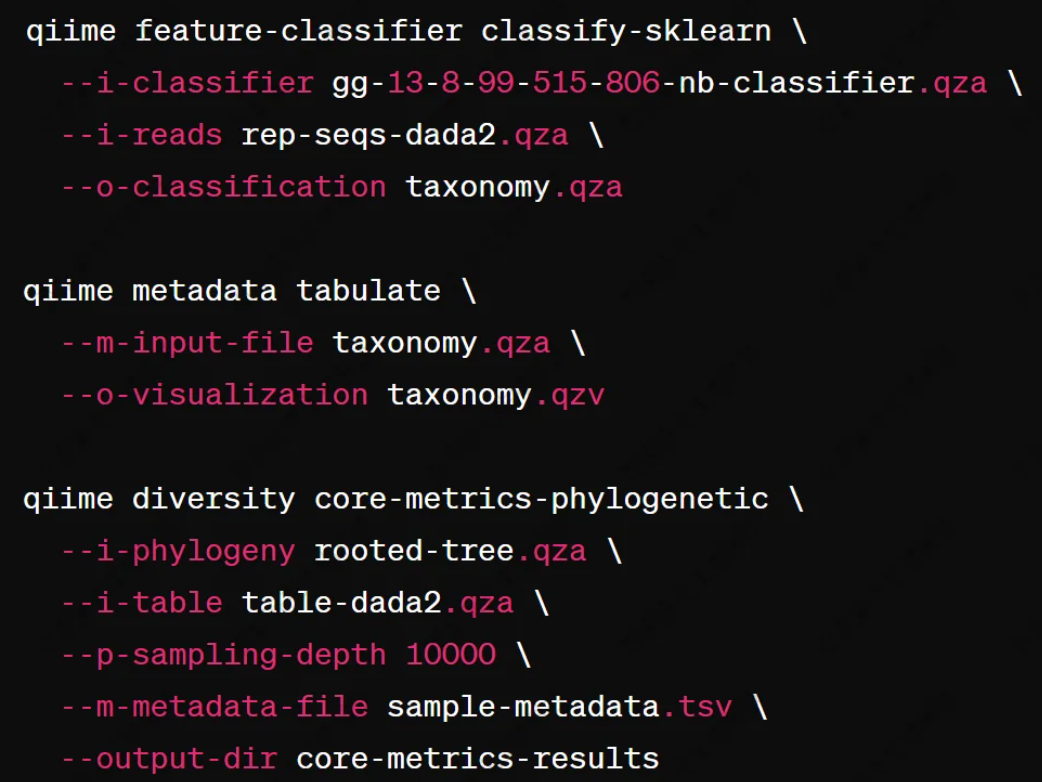
Figure 2
By integrating DADA2 with QIIME2, researchers can achieve high-resolution microbiome profiling using full-length 16S rRNA sequences obtained from third-generation sequencing technologies.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Full-Length 16S/18S/ITS Sequencing Service
How to order?







