16S rRNA Sequencing Bacterial Identification Service
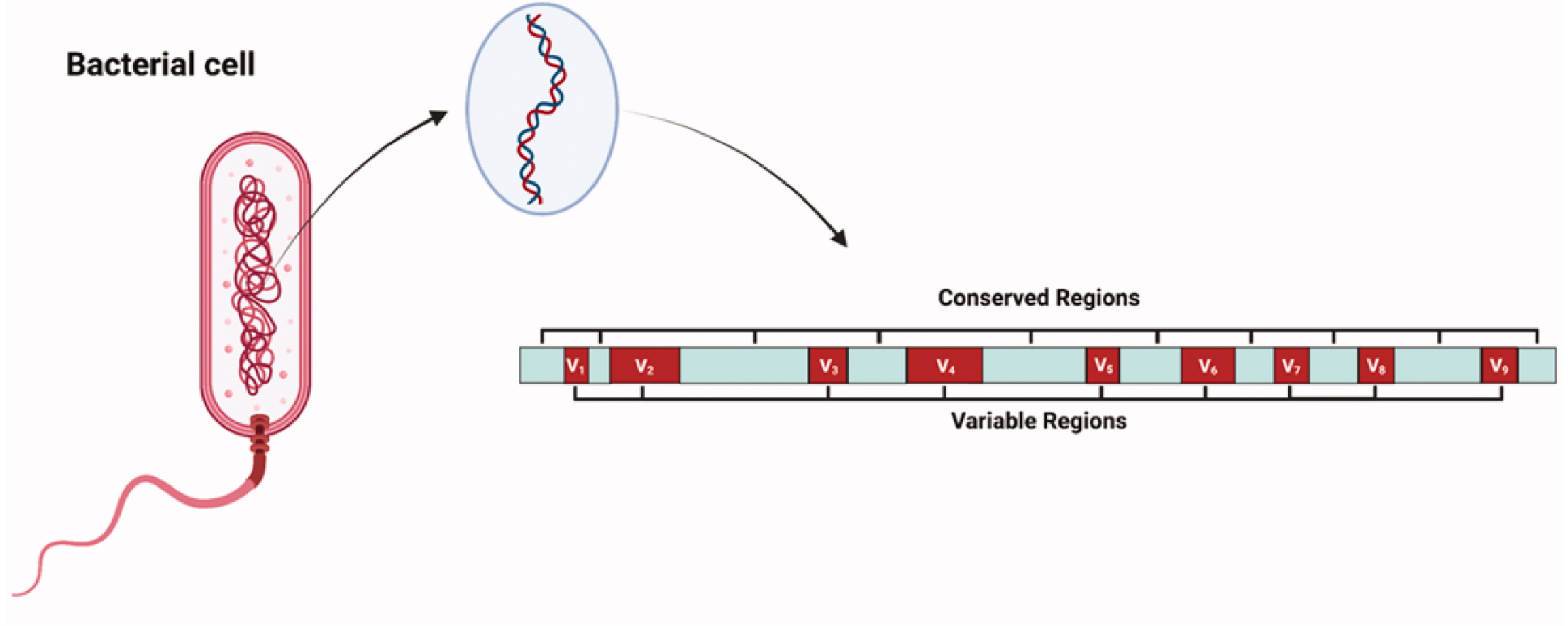
The 16S rRNA Sequencing Bacterial Identification Service is a powerful tool for accurately identifying bacterial species and analyzing microbial communities in diverse environments. The 16S rRNA Sequencing Bacterial Identification Service leverages the unique properties of the 16S rRNA gene, which contains both highly conserved regions, essential for primer binding, and hypervariable regions, critical for distinguishing between bacterial taxa. By targeting these regions, 16S rRNA sequencing enables precise taxonomic classification and phylogenetic analysis, offering unparalleled insights into microbial diversity and bacterial identification.
Lourenco, J. M. et al. ITAL J ANIM SCI. 2022.
Figure 1. Schematic Representation of the 16S rRNA Gene Depicting the Conserved and Variable Regions
The 16S rRNA Sequencing Bacterial Identification Service offers a culture-independent approach to studying bacterial phylogeny and taxonomy, enabling the detection of both culturable and non-culturable species in complex microbiomes. 16S rRNA Sequencing Bacterial Identification Service is particularly useful for analyzing environments or microbial communities that are difficult to study using traditional techniques. It has revolutionized fields such as clinical microbiology, environmental research, food safety, and industrial microbiology, providing researchers with the tools to explore microbial diversity, identify pathogenic bacteria, and gain deeper insights into host-microbe interactions with exceptional precision.
Service at MtoZ Biolabs
Athanasopoulou, K. et al. Biomedicines. 2023.
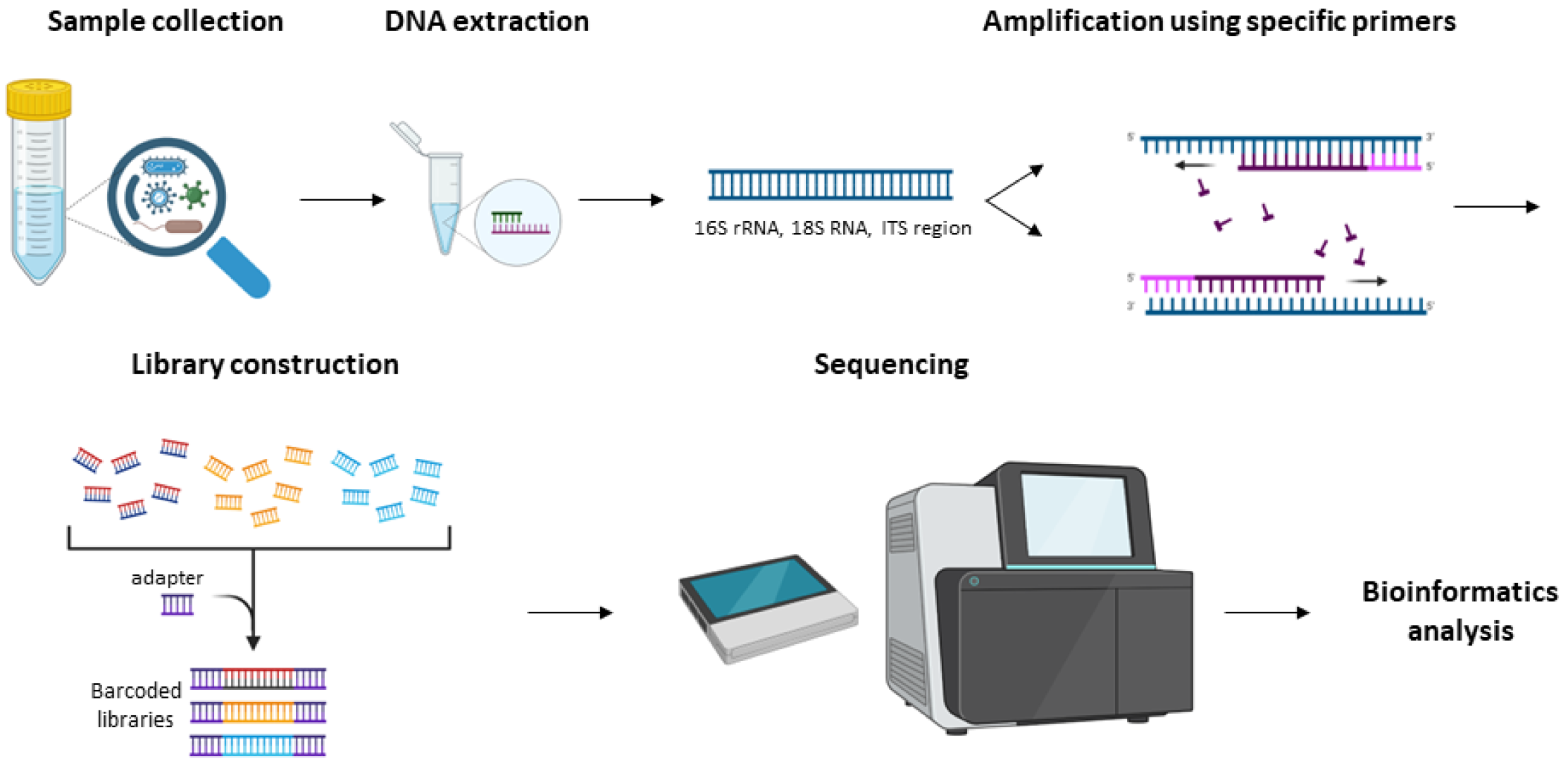
Figure 2. Demonstration of the Workflow for the 16S rRNA Sequencing Bacterial Identification Service
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced 16S rRNA Sequencing Bacterial Identification Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. Customizable Workflows: Offers tailored sequencing strategies to match specific project goals, from primer selection to hypervariable region targeting and data analysis.
3. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
4. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all 16S rRNA Sequencing Bacterial Identification Service data, providing clients with a comprehensive data report.
Applications
1. Clinical Microbiology
Identify pathogenic bacteria in infectious diseases and analyze the human microbiome using the 16S rRNA Sequencing Bacterial Identification Service for health and disease studies.
2. Environmental Microbiology
Investigate microbial diversity in soil, water, and air to study ecological balance, pollution, and biodegradation processes.
3. Food Safety and Quality
Detect foodborne pathogens, monitor spoilage bacteria, and ensure the microbiological safety of food products with the help of 16S rRNA Sequencing Bacterial Identification Service.
4. Industrial Microbiology
Study microbial communities in fermentation processes, bioreactors, and wastewater treatment systems to optimize production and efficiency.
5. Agricultural Microbiology
Analyze plant-associated microbiomes through the 16S rRNA Sequencing Bacterial Identification Service to improve crop resilience, soil health, and agricultural sustainability.
6. Marine Microbiology
Explore microbial ecosystems in aquatic environments to understand biodiversity, nutrient cycles, and potential biotechnological applications.
Case Study
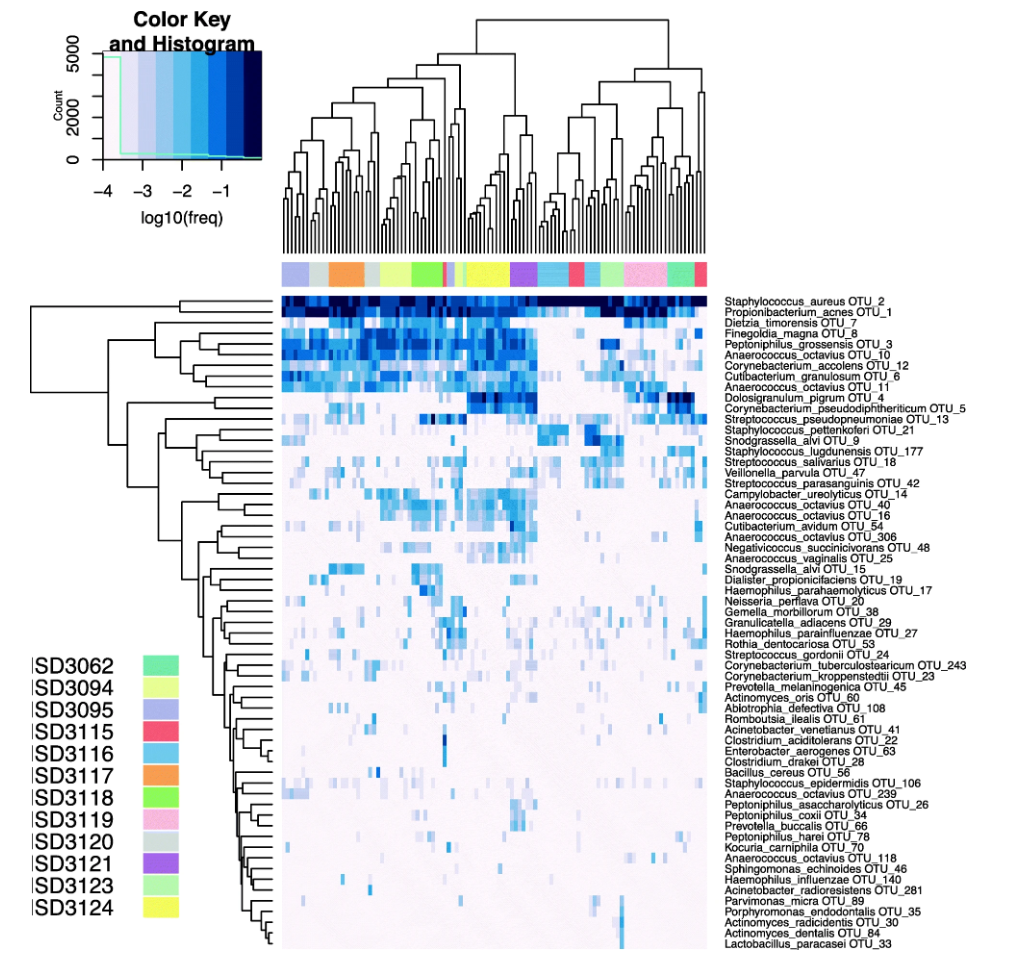
Bacterial Community Profiling of the Healthy Sinonasal Microbiome using 16S rRNA Sequencing Bacterial Identification Service
Earl, J. P. et al. Microbiome. 2018.
Figure 3. Heatmap of Human Sinonasal Microbiome from 12 Subjects
FAQ
Q1: What is the 16S rRNA gene, and why is it used for bacterial identification?
The 16S rRNA gene encodes a component of the 30S small ribosomal subunit, which is essential for protein synthesis in prokaryotes. Its universal presence across all bacterial species, combined with a highly conserved sequence structure, makes it an ideal molecular marker for bacterial identification. The conserved regions facilitate universal primer design, while the hypervariable regions allow differentiation between bacterial species. This dual characteristic ensures both broad applicability and high-resolution taxonomic classification. Compared to traditional culture-based methods, the 16S rRNA Sequencing Bacterial Identification Service enables the identification of both culturable and non-culturable bacteria, providing a comprehensive view of microbial communities in diverse environments.
Q2: What is the role of hypervariable regions in taxonomic classification?
The hypervariable regions (V1–V9) within the 16S rRNA gene contain species-specific nucleotide sequences that allow for fine differentiation among bacteria. During sequencing, these regions are amplified and analyzed to determine taxonomic relationships. Different combinations of hypervariable regions (e.g., V3-V4 or V4-V5) are selected depending on the required taxonomic resolution and the target microbial community. By comparing the sequences of these regions to curated databases, researchers can classify bacteria accurately at the genus or species level. The use of hypervariable regions enhances the precision of the 16S rRNA Sequencing Bacterial Identification Service while maintaining the efficiency of high-throughput workflows.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. The Detailed Information of Bacterial Identification
5. Bioinformatics Analysis
6. Raw Data Files
If you are interested in our 16S rRNA Sequencing Bacterial Identification Service, please feel free to contact us.
How to order?







