GPF-DIA Quantitative Proteomics Services
GPF-DIA Quantitative Proteomics Services leverage Gas-Phase Fractionation (GPF) and Data-Independent Acquisition (DIA) to achieve more sensitive and accurate protein identification and quantification in complex samples. The core principle of GPF-DIA is to use the GPF strategy to perform hierarchical scanning of samples to optimize the data acquisition and analysis process. Unlike traditional DIA methods, GPF-DIA first constructs a high-quality predictive spectral library through multiple narrow-window mass spectrometry scans, which is then used for DIA data analysis. This approach effectively overcomes challenges associated with traditional DIA, including limitations in detecting low-abundance proteins, spectral library dependency, and data interpretation complexity. Leveraging an advanced mass spectrometry platform, MtoZ Biolabs provides comprehensive GPF-DIA Quantitative Proteomics Services covering the entire workflow from sample preparation to data analysis, ensuring high-confidence results for clients.
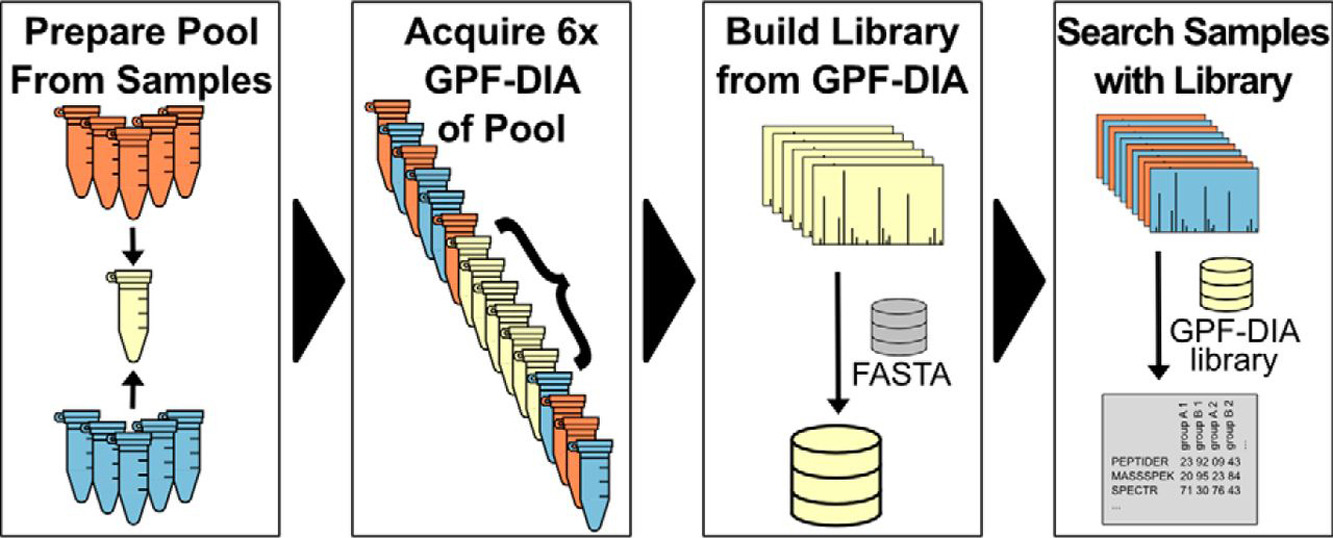
Pino L K. et al. Mol Cell Proteomics. 2020.
Analysis Workflow
1. Sample Preparation
Proteins are extracted and digested using a standardized workflow. Peptides are purified via high-performance liquid chromatography (HPLC) to remove impurities and enhance the accuracy of mass spectrometry detection.
2. GPF-DIA Data Acquisition
High-resolution mass spectrometry is used to perform fractionated scanning across multiple m/z windows, acquiring high-resolution data-dependent acquisition (DDA) or narrow-window DIA data to ensure high-quality spectral information.
3. Predictive Spectral Library Construction
High-quality virtual spectral libraries are generated using AI-based predictive models such as Prosit and DeepMass.
4. DIA Data Acquisition
Standard DIA methods are employed to perform wide-window scans on experimental samples, ensuring high-throughput data acquisition.
5. Data Analysis and Quantification
Advanced software such as DIA-NN, Spectronaut, and Skyline is utilized for DIA data analysis based on the predictive spectral library. A comprehensive data report is provided, including protein quantification results, differential expression analysis, and functional enrichment analysis.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs offers GPF-DIA Quantitative Proteomics Services to support high-sensitivity, high-precision quantitative large-scale proteomics analysis. This service is suitable for various research fields. Contact us to customize your tailored research solution.
Applications
GPF-DIA Quantitative Proteomics has wide applications across various research fields, including:
Disease Biomarker Research
GPF-DIA Quantitative Proteomics enables the identification of disease-associated proteins and enhances the detection of low-abundance proteins.
Systems Biology
Helps analyze protein interaction networks and signaling pathways to uncover dynamic regulatory mechanisms within biological systems.
Precision Medicine & Translational Research
GPF-DIA Quantitative Proteomics can be used to accurately quantify protein expression levels for personalized medicine research, such as cancer proteomics and immunotherapy response prediction.
Drug Discovery
Supports drug target screening and mechanism of action studies by delivering high-resolution quantitative proteomics data, facilitating new drug development.
FAQ
Q. How can the window segmentation in GPF-DIA Quantitative Proteomics be optimized to ensure maximum coverage?
Optimizing window segmentation in GPF-DIA requires a balanced consideration of mass spectrometry resolution, sample complexity, and experimental objectives. For highly complex samples (e.g., plasma or tissue), a gas-phase fractionation strategy with 6-10 m/z windows is typically recommended to reduce ion competition and improve low-abundance peptide identification rates. The selection of window width should ensure an even distribution of peptides within each window, avoiding overly narrow windows that lead to data redundancy or excessively wide windows that compromise resolution. Additionally, machine learning-based predictions (such as DIA-NN and Prosit) can be integrated to dynamically optimize window segmentation, further enhancing data quality and protein identification depth.
Q. How can DIA data analysis strategies be optimized to reduce false matches?
The key to optimizing DIA data analysis strategy lies in high-quality library construction, fragment ion matching strategy and FDR control:
1. High-Quality Spectral Library
Leveraging deep-learning-based predicted spectral libraries (e.g., Prosit) instead of traditional DDA-based libraries helps eliminate experimental biases and improve matching accuracy.
2. Optimized Fragment Matching
Use advanced algorithms such as DIA-NN and Spectronaut, combined with machine learning methods to optimize fragment matching and improve the peptide identification rate.
3. FDR Control
Implementing a 1% protein-level FDR and applying stricter q-value filtering at the peptide level minimizes false-positive identifications, particularly for low-abundance peptides.
4. Internal Standard Calibration
Use iRT peptides or heavy labeled proteins for calibration to improve the stability and reliability of quantitative data.
By applying these optimization strategies, DIA data analysis accuracy can be significantly improved, reducing false matches and ensuring high-confidence results.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study used GPF-DIA Quantitative Proteomics to optimize DIA phosphoproteomics, increasing the number of phosphoprotein identifications, the sensitivity of low-abundance site detection, and quantitative reproducibility, and combined with deep learning prediction spectral libraries to further optimize data analysis. This has expanded the application of DIA in phosphoprotein research and provided new ideas for future high-throughput, precise quantitative phosphoproteomics analysis.
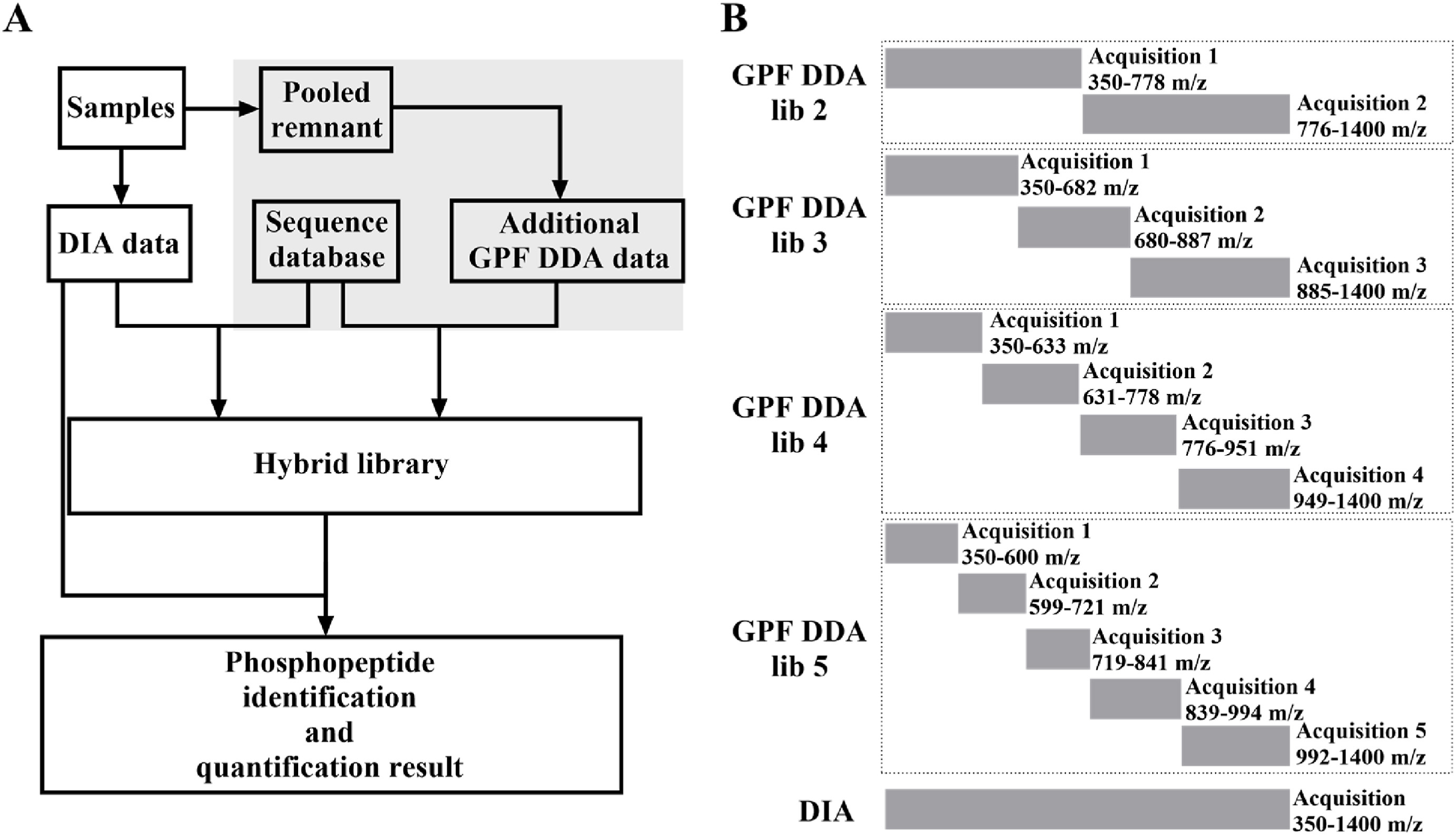
Tu Z W. et al. Heliyon. 2025.
How to order?







