FACS Based Exosome Characterization Service
FACS Based Exosome Characterization Service uses the fluorescence-activated cell sorting (FACS) platform to perform multiplex fluorescence labeling analysis of specific protein markers on the exosomal surface, enabling multi-level characterization of exosomes, including subpopulation typing, purity assessment, and surface protein quantification. Exosomes are nanoscale extracellular vesicles derived from a variety of cells, widely present in bodily fluids such as blood, urine, cerebrospinal fluid, and more. They play key roles in intercellular communication, disease development, and immune regulation. Due to their high heterogeneity and significant functional differences between exosomes from different sources, precise characterization, especially the high-sensitivity and high-resolution detection of surface markers, is critical for disease research and precision medicine. The FACS based exosome characterization technique allows for efficient, multi-marker analysis of individual exosomes, making it one of the high-resolution technologies used in current exosome research. Through the FACS platform, MtoZ Biolabs offers the FACS Based Exosome Characterization Service providing precise identification, quantification, and classification of exosomal surface markers, enabling exosome classification, source tracking, and disease relevance analysis.
Analysis Workflow
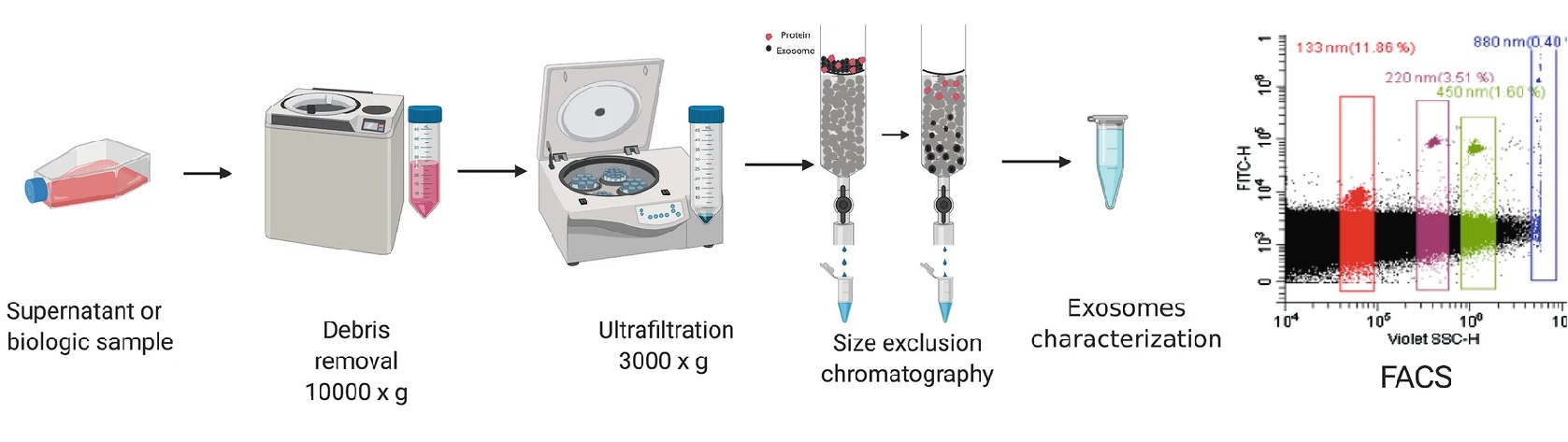
Cynthia López-Pacheco, et al. Analysis of Tumor-Derived Exosomes by Nanoscale Flow Cytometry. 2020.
1. Sample Preparation
Isolate exosomes using standard methods. Perform pre-testing of particle size and concentration to ensure the exosome samples meet the requirements for flow cytometry analysis.
2. Fluorescent Labeling
Select fluorescently conjugated antibodies for target surface proteins and incubate for specific labeling. Set up single staining and negative controls to determine the specificity of the fluorescent signal.
3. Flow Cytometry Analysis
Analyze the labeled exosome samples using FACS. Collect fluorescence and scattering signals, ensuring single-particle resolution and avoiding aggregation errors.
4. Data Analysis
Use specialized flow cytometry data analysis software to process the data. Differentiate exosome subpopulations, calculate the expression frequency and intensity of surface markers, and provide various visual results such as exosome distribution maps and fluorescence intensity histograms.
Applications
Disease Diagnosis and Prognosis Evaluation:
Exosomal surface markers can serve as biomarkers for early disease diagnosis, classification, and prognosis assessment (such as in cancer, neurodegenerative diseases, and immune disorders).
Tumor-Derived Exosome Detection:
FACS Based Exosome Characterization Service can accurately identify tumor-derived exosomes, such as those expressing EpCAM, HER2, and PD-L1, aiding in non-invasive liquid biopsy.
Immune Exosome Profiling:
Analyze exosomes secreted by immune cells to study their roles in immune regulation and inflammatory responses.
Drug Delivery Exosome Development:
Identify surface characteristics of engineered exosomes used in drug delivery systems, ensuring targeting and biocompatibility.
Basic Research:
Explore the roles of exosomes in cell communication and disease progression, revealing their functional mechanisms.
Services at MtoZ Biolabs
Based on advanced experimental platforms, MtoZ Biolabs not only offers the FACS Based Exosome Characterization Service but also provides in-depth exosome content analysis in combination with proteomics or RNA sequencing. Additionally, we offer one-stop integrated solutions for comprehensive exosome analysis. MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption.
FAQ
Q. How to Choose the Appropriate Surface Markers to Specifically Label Exosomes?
When selecting surface markers for exosomes, membrane proteins associated with exosome biogenesis and release are commonly chosen. Well-known markers include CD63, CD9, and CD81, which are typically considered classic exosomal markers. However, studies have shown that these markers are not exclusive and may not fully represent all exosome subpopulations. Exosomes from different sources (e.g., cancer cell-derived vs. normal cell-derived exosomes) may express different markers. Some exosomes may lack these classic markers or exhibit other surface proteins (e.g., EpCAM, Integrins), which may not be recognized by traditional markers.
To comprehensively characterize the heterogeneity of exosomes, it is recommended to combine multiplex marker analysis using multiple specific antibodies, especially those targeting markers of exosome subpopulations derived from different cell types.
Case Study
This study utilized FACS based exosome characterization technology to analyze the composition of exosome subpopulations in metastatic breast cancer patients and their relationship with the lymphocyte ratio. The results revealed that exosome subpopulations play a significant role in tumor immune evasion, tumor metastasis, and other processes, while the lymphocyte ratio may influence the progression of these processes.
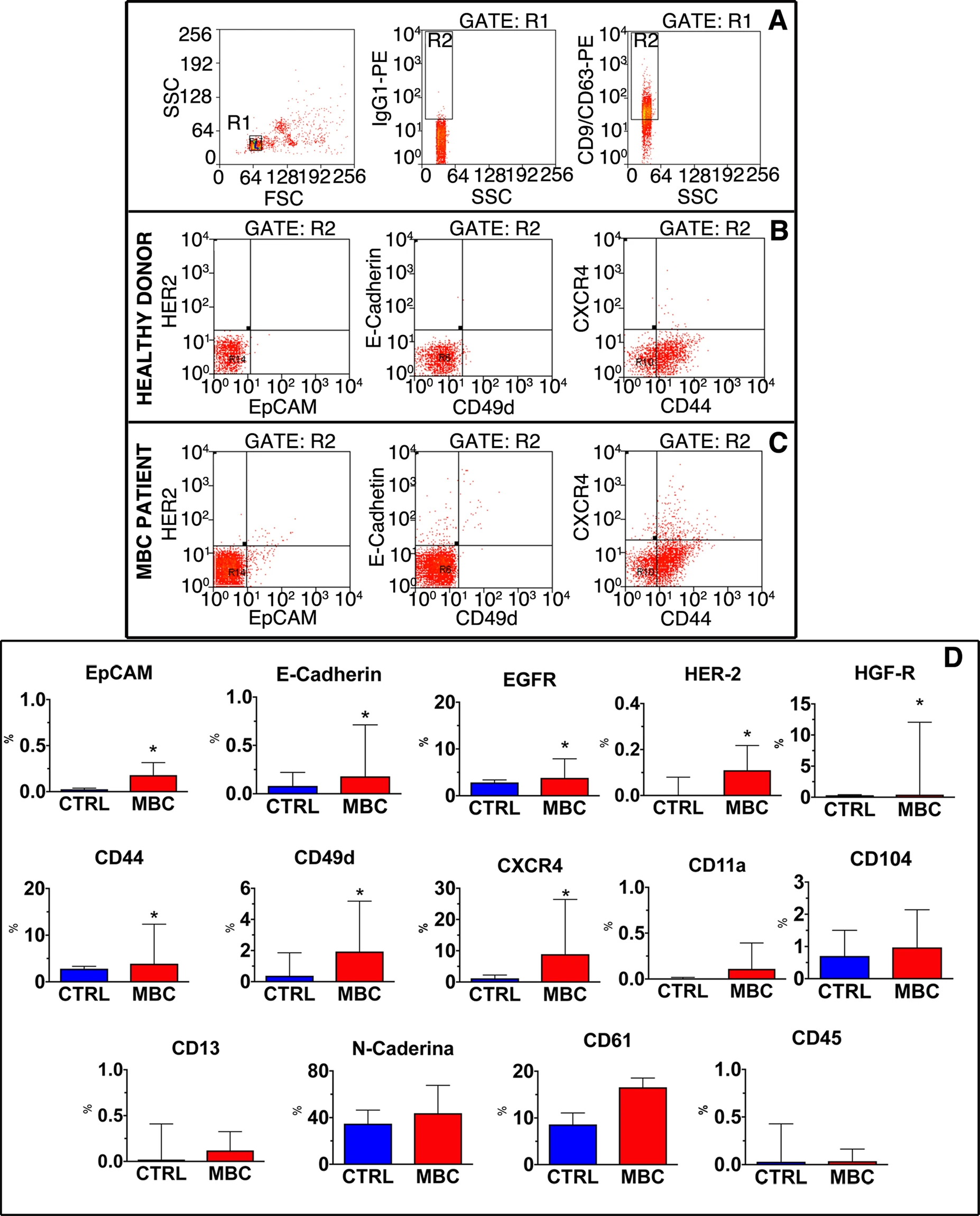
Gerratana L. et al. Scientific Reports. 2020.
How to order?







