Exosomal lncRNA Sequencing Service
Exosomal lncRNA Sequencing Service utilizes high-throughput sequencing technology to comprehensively identify and quantify the types, expression levels, and potential regulatory roles of long non-coding RNAs (lncRNAs) in exosomes, revealing their molecular mechanisms in physiological processes or disease development. LncRNAs are non-coding RNAs longer than 200 nucleotides (nt) and are widely found in eukaryotes. Unlike mRNAs, lncRNAs do not encode proteins, but they play crucial regulatory roles within cells, such as transcriptional regulation, cell process regulation, and post-translational regulation. Exosomes contain abundant lncRNAs, which are encapsulated to protect them from external environmental influences and can be transferred between cells to regulate gene expression and function in distant target cells.
In recent years, exosomal lncRNAs have been found to be widely involved in gene regulation, signal pathway modulation, and tumor microenvironment shaping, with significant potential as biomarkers and therapeutic targets in diseases such as cancer and cardiovascular disorders. Studying exosomal lncRNAs helps improve the understanding of the molecular mechanisms underlying disease development and provides new approaches for clinical diagnosis and treatment.
Based on high-throughput sequencing platforms, MtoZ Biolabs provides Exosomal lncRNA Sequencing Service to systematically sequence and analyze exosomal lncRNAs, offering precise molecular data support for both basic research and clinical translational studies.
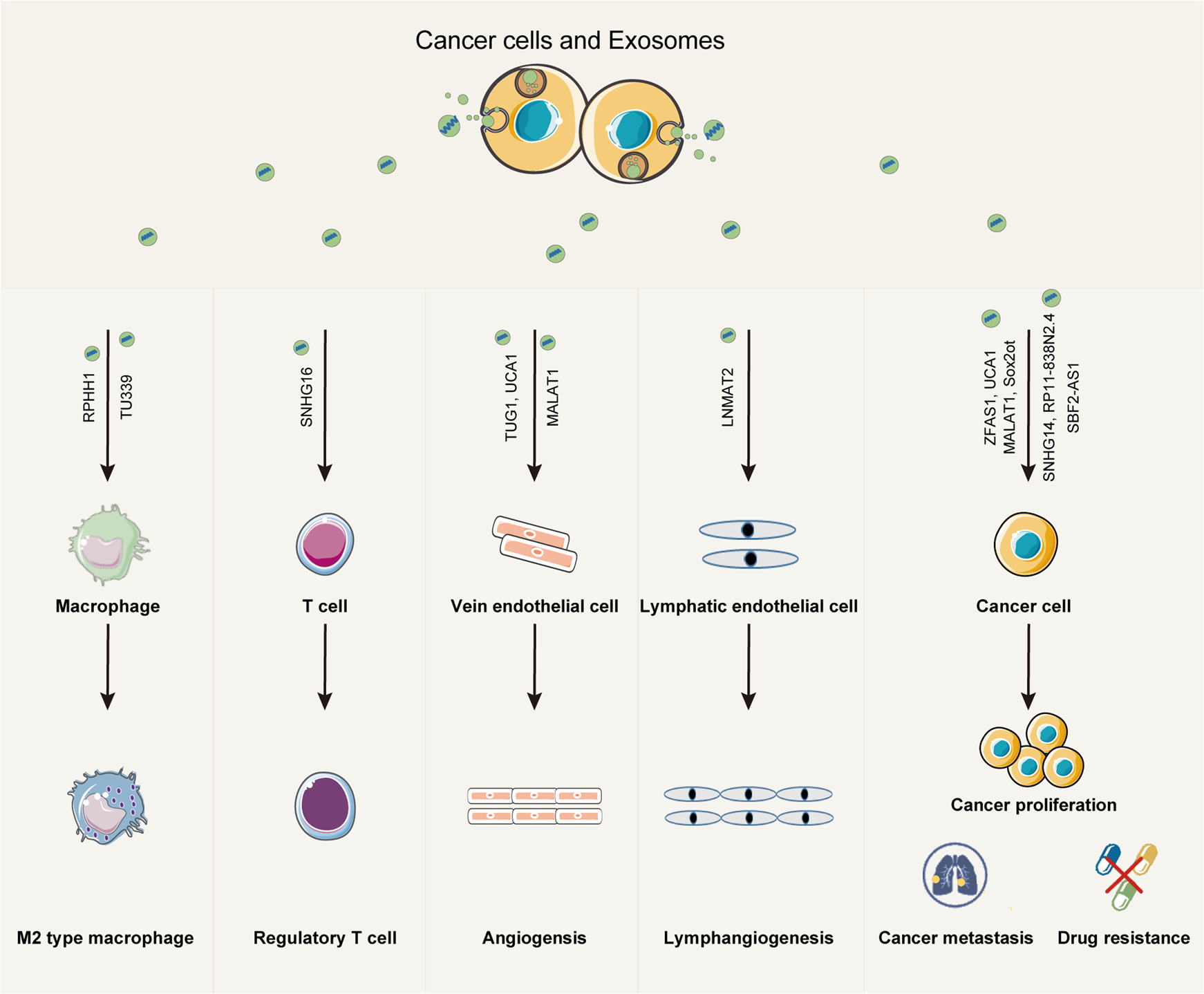
Zhang W. et al. Experimental & Molecular Medicine. 2021.
Figure 1. The role of exosomal lncRNAs in cancer biology.
Analysis Workflow
1. Exosome Isolation: Isolate high-purity exosomes from the sample.
2. RNA Extraction: Efficiently extract total RNA from exosomes.
3. rRNA Removal and lncRNA Enrichment: Remove ribosomal RNA and enrich for lncRNA.
4. Library Construction: Fragment lncRNAs, add adapters, reverse transcribe to cDNA, and amplify via PCR to construct high-quality sequencing libraries.
5. High-Throughput Sequencing: Perform paired-end sequencing using platforms such as Illumina to achieve comprehensive coverage of lncRNA sequences.
6. Data Analysis and Reporting: Conduct data analysis and bioinformatics analysis, providing a detailed report.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Exosomal lncRNA Sequencing Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Exosomal lncRNA Sequencing Service data, providing clients with a comprehensive data report.
4. Comprehensiveness: Cover both known and novel lncRNAs to construct a complete expression profile.
5. High Resolution: Paired-end sequencing for precise revelation of lncRNA structures and splice variants.
FAQ
Q. How much Sample is Required for Exosomal lncRNA Sequencing Service?
The required sample amount depends on the sample type and the expected exosome yield. It is recommended to provide at least 200–500 µL of plasma/serum or 10 mL of cell culture supernatant per sample. For special samples, customized solutions can be provided, and specific requirements can be discussed and confirmed based on the project needs.
Q. What Information can be Obtained from the Sequencing Results?
You will receive a detailed comprehensive report, including:
1. Comprehensive experimental details
2. Materials, instruments, and methods
3. Identification and annotation table of known and novel lncRNAs
4. lncRNA expression profiles and quantification data
5. Differential expression analysis
6. Bioinformatics analysis
7. Raw data files
Case Study
This study utilized exosomal lncRNA sequencing technology to systematically analyze the exosomal lncRNA expression profile in an acute myocardial ischemia (AMI) model and identified HCG15 as a significantly upregulated exosomal lncRNA. The research found that HCG15 promotes myocardial cell inflammation and apoptosis by activating the NF-κB/p65 and p38 MAPK signaling pathways, exacerbating ischemic injury. In contrast, inhibiting or removing HCG15 significantly alleviated myocardial damage. These findings suggest that exosomal lncRNA HCG15 is an important regulatory factor in myocardial ischemic injury and could serve as a new target for intervening in AMI.
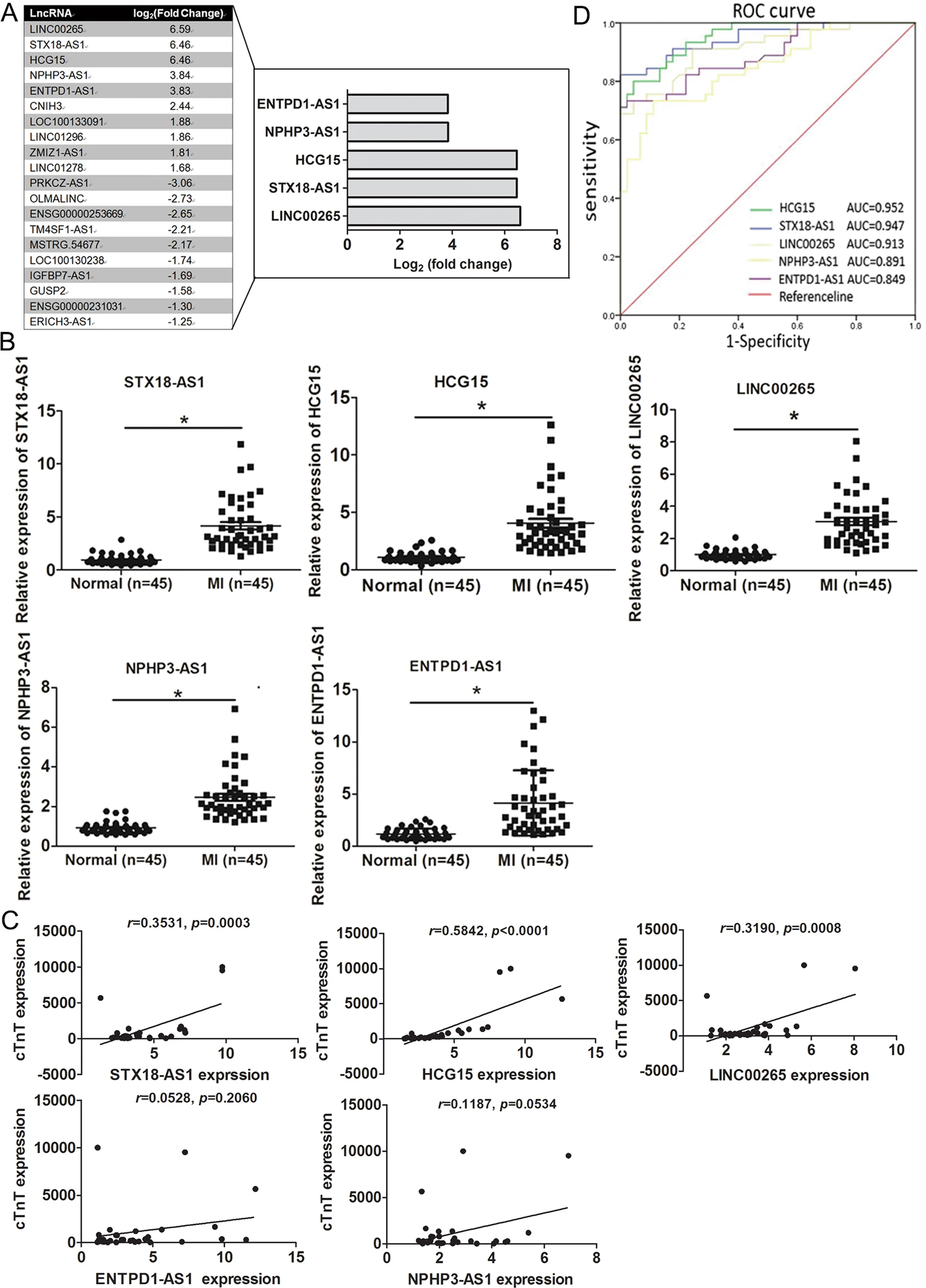
Lin B. et al. Cell Death & Disease. 2021.
Figure 2. Identification of the differentially expressed exosomal lncRNAs from the serum of AMI patients and healthy controls.
How to order?







